Axial - DiCE Molecules
Surveying great inventors and businesses
Axial invests and partners in early-stage life sciences companies. If you or someone you know has a great idea or company in life sciences, Axial would be excited to get to know you and possibly invest in your vision and company — info@axialvc.com
To simplify this newsletter, if you want to receive:
Early ideas around life sciences technologies and businesses, sign up for https://axialobservations.substack.com/
New financing events with some analysis, sign up for https://axialfc.substack.com/
Analysis of life sciences inventors and their inventions, sign up for https://axialinventors.substack.com/
Reviews of life sciences decks, sign up for https://axialdeck.substack.com/

DiCE Molecules is pushing the business model possibilities for small molecule companies. Centered around a directed evolution technology for DNA-encoded libraries, DiCE is a useful case study on how hard it is to build a licensing business for small molecules and how to use a small molecule platform to find rare chemical matter to translate into valuable products.
Founded in 2013 by Kevin Judice, Phil Patten, and John Bedbrook, DiCE was premised on bringing directed evolution to chemistry. The founders took work out of the Harbury Lab at Stanford to combine mesofluidic technologies with directed evolution to build a DNA-encoded library with both screening and optimization built-in, which the company calls directed chemical evolution or DiCE.
In the 1990s, Kevin and John as well as Pehr Harbury were postdocs at UC Berkeley in the Schultz Lab (a pioneer in chemical biology). Harbury became a professor in the Stanford biochemistry department doing work in combinatorial chemistry and directed chemical evolution. While Judice went on to work as a scientist at Genentech then moving up as a senior director there and afterwards founding Achaogen to develop new antibiotics, a company worth its own case study, and Cidara Therapeutics to treat fungal infections. Patten was a VP at Maxygen (protein engineering via DNA shuffling), a SVP at Achaogen, and then a fellow at Pioneer HiBred. Bedbrook was president of agriculture at Maxygen and sold his company Verdia that was ultimately bought by Pioneer. DiCE was really a company that took 20 years of relationship building to found.
In 2013, the idea of DiCE formed when Harbury began having discussions with Patten and Judice - a reunion of sorts from their postdoc days. Bedbrook was brought in for his experience, and the company set off to build a business that brought directed chemical evolution to small molecule drug development. At the time incumbents like HitGen and X-Chem had brought DNA-encoded libraries to the market. DiCE wanted to bring directed evolution to DNA-encoded libraries.
DiCE’s platform is centered around a methodology that almost acts as a router for small molecules to a given target. The platform selects/optimizes large libraries of small molecules (100M-1B molecules) against high value targets (i.e. undrugged proteins, PPIs). Companies like HitGen were doing the same, but DiCE did something unique by both screening and selecting for small molecules in each screen. This enabled DiCE to search the full landscape of structure-activity relationships (SAR) for a given target. Usually during a screen for a DNA-encoded library, one would find a positive SAR cluster and characterize the chemical matter manually for potency, selectivity, and other drug-like properties. DiCE automated this process by collapsing it into one experiment and saving years of work. DNA-encoded libraries sped up the hit generation process; screening a larger set of molecules against targets increases the odds of getting a potential drug lead. DiCE took this a step further by also speeding up the hit-to-lead process by automating optimization within its DNA-encoded library platform.
With this unique set of technologies, DiCE was able to pursue a self-financing model and build a hybrid business model between licensing and internal development. In 2016, DiCE was able to partner with Sanofi to use its platform against 12 targets for a little over $50M in up-front payments and an equity investment as well as downstream incentives. This helped scale up DiCE and set the company up to build its own pipeline in a less dilutive way. DiCE was a company that took two decades of work before it could get started, and now it is a valuable case study on how to use a set of inventions to get less dilutive financing and build a unique business model.
Key findings
For DiCE and DEL companies in general, the key levers for their platforms are the library size (millions to billions of molecules), which properties to select for especially beyond just target affinity, how many cycles required to get a hit/lead, and what types of medicinal chemistry will be required to take the hit/lead and make it into a drug.
So how many customers are there out there that want to buy a DEL? Conservatively the number is in the hundreds - a good estimate is the number of customers Veeva has: a little over 600 companies. The upper limit for revenue from selling DEL libraries is somewhere between $60M to $120M for a given target. It’s probably on the lower end because not every mid-to-large biopharma company has small molecule programs. For multiple targets, this easily becomes a billion dollar opportunity.
What ultimately happened was that DiCE decided to focus on an internal pipeline with their Sanofi partnership still active. DiCE can always revisit licensing out more mature assets; however, the market opportunity for DELs was on the path to commoditization compelling the company to develop its own medicines. Antibody licensing businesses have had more attractive economics on their deals because they have a history of better clinical outcomes. DELs don’t have that track record just yet. Maybe DiCE Molecules becomes DiCE Medicines.
DiCE was founded with the mission to transform small molecule discovery. They brought a new feature, directed evolution, to a validated technology, DELs. DiCE used this to strike up a unique deal that financed a lot of their internal development at very favorable, non-dilutive terms. DiCE is a case study on how a licensing model can be an efficient, both scientifically and financially, way to build out an internal pipeline. The next 7 years of DiCE will be driven by the ability of their DELs to find rare chemical matter that have better odds to become successful medicines with the goal of replacing approved biologics with a small molecule.
Technology
DNA-encoded libraries (DEL) tag each compound with a unique DNA barcode (~20-base-pair DNA sequences) to enable templated synthesis and tracking. The idea of a DEL emerged in the early 1990s, when the DiCE founders were postdocs, from Scripps - https://www.pnas.org/content/89/12/5381 The premise of DELs was driven by the difference between small molecules and biological molecules: the latter are produced by a genetic code and the former are not. Biological molecules can replicate and be selected for; whereas, small molecules don’t have these intrinsic features. The idea of a DEL started with the idea that adding a piece of DNA to a small molecule can imbue a code onto them and allowing the application of selective forces to identify small molecules with certain features. When DiCE was founded, the first wave of DEL companies had hit:
Version 1: large libraries of small molecules with little-to-no directed evolution; companies like X-Chem, HitGen, and Vipergen; bottleneck was sequencing capabilities
Version 2: larger libraries using directed evolution; companies like DiCE, Ensemble (from the Liu Lab), and Nuevolution (now part of Amgen); sequencing became cheaper and more powerful
Version 3 will have to focus on diversity, medicinal chemistry, and relieving the bottlenecks from pooling and designing new selection methods; sequencing is pretty cheap with new tools like machine learning and microfluidics coming in. Machine learning will be particularly useful to map out SARs more rapidly. An exciting development is screening libraries against whole cell environments and membranes going beyond soluble protein targets.
Initially, the size of DNA-encoded libraries were relatively small and the compounds within them were often large and lipophilic (not good for drug candidates). The first generation of companies like HitGen had to develop new water-based chemistry to improve the diversity of their libraries and generate more building blocks to increase library size. As sequencing costs came down in the 2000s, more hits were able to be screened and discovered. This set up the second generation to bring directed evolution to DNA-encoded libraries - it now costed a few hundred bucks to sequence up to a million hits.
Medicinal chemistry will always play a role in this field; DNA-encoded libraries are a complementary tool to methods like high-throughput screening, fragment-based design, and displays. The ability to perform things like aminations and Wittig reactions will always be required in drug development. DELs enable rapid screening of billions of compounds. What made DiCE unique was that its technology pushed out the need for a medicinal chemist from lead optimization to candidate development. This got Sanofi really excited to execute a deal with DiCE.
DELs are centered around combining DNA barcoding with combinatorial chemistry to synthesize and screen small molecules up to the size of macrocycles/peptides. The main advantage is the ability to screen a library of a billion compounds in a single test tube where hits (and their synthesis route) can be identified through DNA sequencing.
The two main methods to construct DELs are split and pool and one pot synthesis. DiCE’s platform relies on a certain version of split and pool. In split and pool, DNA-barcoded small molecules (s) are pooled together and split into separate groups (g) for the synthesis reaction and repooled - this enables the library to grow by (sxg)^r after each reaction (r). In one-pot synthesis, the DNA barcodes act as templates for small molecule synthesis where the tagging and reaction occur in the same vessel. Split and pool enables larger and larger libraries, while a one-pot synthesis method enables new types of reactions that do not normally occur in solution at low concentrations.
The Liu Lab (Harvard, formed the basis of Ensemble) was the inventor of one-pot/DNA-templated synthesis for DELs. Whereas, the inventions that formed the basis of DiCE was from the Harbury Lab, which pioneered programmed split and pool methods. The simple split and pool approach is just tagging all the compounds for identification. The method DiCE relies on has a little more complexity in the library set up. Programmed split and pool uses the barcode not only to identify the molecule but select for certain properties - an array of oligonucleotides are used to separate the barcodes. A mixture of the templated barcodes are flowed over an array of oligonucleotides in a fluidic device - this separates each barcode according to the oligo it hybridizes to. After this, the chemical compounds are added to link to their corresponding barcodes and are pooled together. The oligo array is really important because it enables the barcode to be amplified during the screening step. The splitting process using the array of oligos is started again; this is done iteratively - https://www.ncbi.nlm.nih.gov/pmc/articles/PMC434148/
Across these three methods for DELs, their advantages and problems are:
One-pot synthesis
Advantages: Enables more complex reactions to occur
Problems: Have to design the barcodes very carefully to make sure they don’t self-hybridize
Programmed split and pool
Advantages: Each round enriches for a higher concentrations of hits and during the array step and chemical compounds can be added in a wide range of solvents beyond water
Problems: More complex set up using the array of oligos, which requires higher input costs and a fluidic device
Tagged split and pool
Advantages: Through iterative steps of pooling and splitting, large small molecule libraries can be created from a simple starting point of chemical compounds
Problems: Valuable molecules can be lost from detection during the pooling step and there is no way to optimize for molecules during the screening process other than maybe binding affinity
The first step for all these methods all require adding a chemical compound to a piece of DNA. This reaction often occurs in water, but for DiCE’s method they can use other solvents, which enables them to build unique library types. During a DEL screening, hits can be found but then resynthesizing them can be a pain especially if the chemical compounds used in the beginning are rare or are hard to make in large amounts. This is a major issue that DiCE was able to avoid; compared to their competitors, DiCE had a wider menu in the starting compounds they could use, which helped them not run into the problems companies like Ensemble faced. Overview of synthesizing a DEL with a split and pool method:
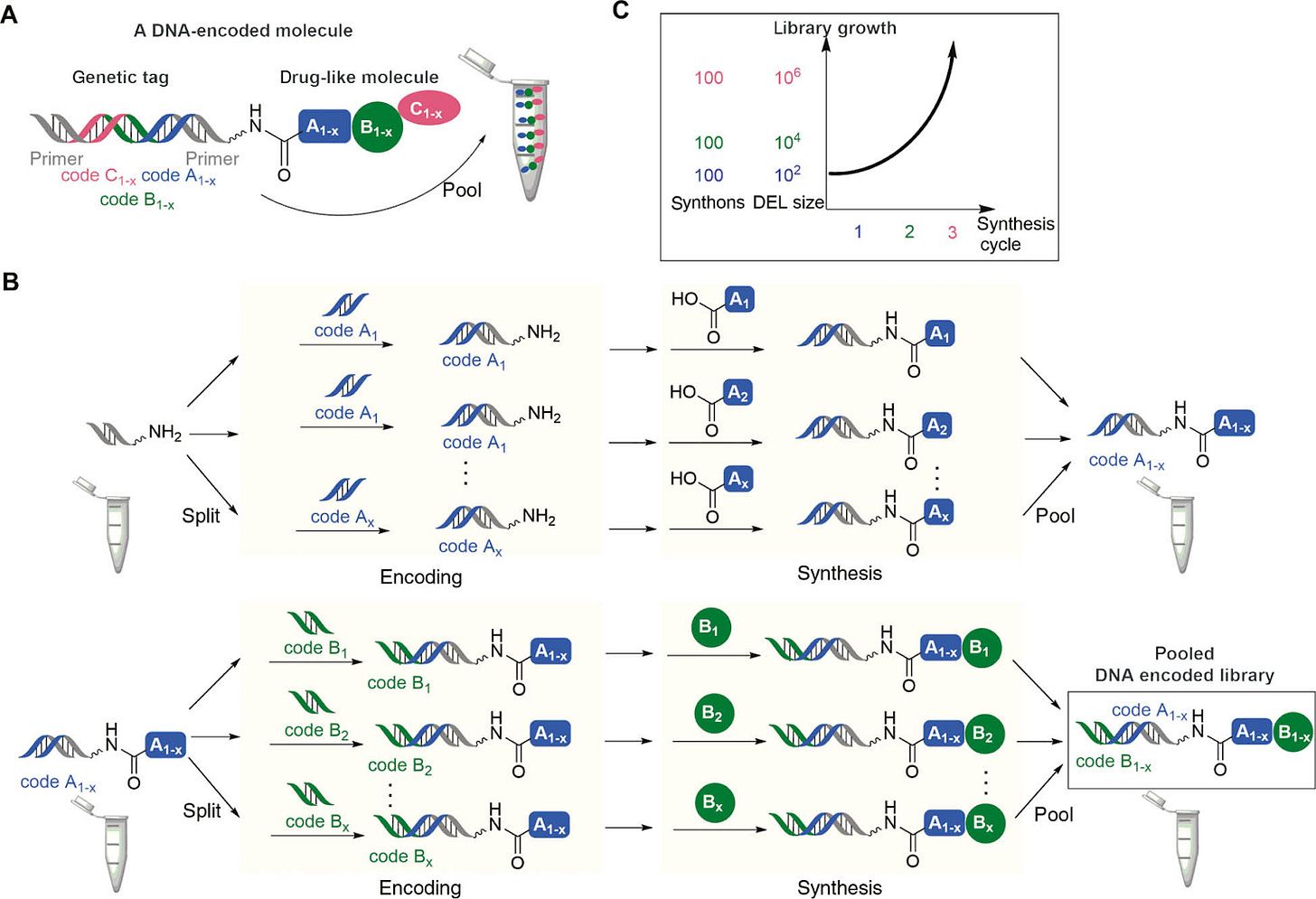
Split and pool method to make a DEL. Source: Degruyter
After the library is built, the DEL is screened against a target. This can be done in a single tube where the target protein is incubated with the DEL. Often the target protein is immobilized and soluble. You can also add a DNA barcode to the target itself, which will hybridize with the barcode on any hits (this requires a secondary primer extension step). You can also photocrosslink a hit and a target. Also, a barcoded target and compound can be emulsified in a micelle if they interact where the target barcode is ligated to the compound barcode. When compared to high-throughput screening (HTS), a DEL screen has major advantages in terms of scale, speed, and cost. In HTS to screen a million compounds against 10 targets requires 10 million separate experiments. Whereas for a DEL, doing the same screen requires 10 experiments - one for each target.
Other advantages of DELs versus other drug screening methods are:
DELs can store hundreds of millions of molecules in a tube, that means less need for freezers and compound management infrastructure
DEL screens can be done on a lab bench again meaning lower infrastructure needs and the ability to run multiple screens pretty easily
DEL screens only need a few micrograms of a target protein, which means lower reagent costs per screen
However, DELs also have several issues. Many of the DEL methods described can only use water-based chemistry. DiCE solved this by using an oligo array that is attached to paper, which allows them to use different solvents. Also, the DNA barcode attached to the compound could interfere with how the compound interacts with the target. Moreover, the screening reaction conditions (upper limit of ~200 °C, need a solution with at least 20% water) are limited to ones that do not damage DNA; DNA that is damaged cannot get sequenced. Other issues are around prioritizing hits and resynthesizing them. The former is an issue found across all modalities: you get 20-200 hits from a screen, pick 1-3 as drug leads? For DELs, the latter depends on the initial chemical compounds used to build the library. DiCE gets around the choosing problem by adding selective forces to its screening process and solves the resynthesis problem by having a large set of chemical compounds they can start off with.

Source: Biopharmatrend
For DiCE, a DEL is screened against a target selecting for features like affinity. After a wash step, the hits are amplified via PCR with their molecular structures decoded through sequencing. Between rounds, recombinations between barcoded molecules creates diversification and effectively creates a much larger library than the initial DEL. Also, in between rounds, DiCE restores the molecules within the library to their original ligand concentrations to more easily select for drug-like features (i.e. logD, Cyp stability). DiCE iterates across this process again and again to find drug leads. This process helps DiCE explore SARs more efficiently by automating the affinity maturation step to find leads that might have low odds of actually existing in the initial DEL population. This could only really be done with a programmed split and pool method and allows DiCE to play around with a wider set of selection criteria even things like phenotypic screening (i.e. allosteric sites, E3 ligases). In short, DiCE uses the DNA barcode to both label compounds and provide their synthesis instructions - this creates a DEL that has small molecules with heritable features.
The three main advantages DiCE have are:
Affinity maturation of small molecules during the diversification step in the screening process. This also allows for multiple rounds of selection for many drug-like properties including affinity, selectivity, metabolic stability, and cell permeability.
Their process to diversify the library and reset ligand concentrations after each round of screening helps DiCE explore more of the SAR landscape in one experiment. This is a major automation step that saves years of work through the production of drug leads.
DiCE produced a custom fluidic device, which they call a mesofluidic ribosome that allows the company to use programmed split and pool DEL methods. Think of this device as a DNA router.
For DiCE and DEL companies in general, the key levers for their platforms are the library size (millions to billions of molecules), which properties to select for especially beyond just target affinity, how many cycles required to get a hit/lead, and what types of medicinal chemistry will be required to take the hit/lead and make it into a drug.

Market
DiCE’s market can be a little complex. How do you value the market of a library of small molecules? It’s straightforward to size the opportunity for a drug in specific diseases: patient population x drug cost. However, how much revenue can be built out from selling small molecule libraries and getting royalties as well? At the time of DiCE’s founding, HitGen and other comparables were selling libraries for $100K-$200K per target with some milestone payments and royalties baked in. At the lower-end, a DEL can be bought for $12K. Overall, this pricing point is a major value proposition to potential customers. The cost of a HTS for a million molecules costs a few million dollars. The ability to screen the same amount of chemical matter for 10x lower the costs is pretty compelling.
So how many customers are out there that want to buy a DEL? Conservatively the number is in the hundreds - a good estimate is the number of customers Veeva has: a little over 600 companies. Therefore, the upper limit for revenue from selling DEL libraries is somewhere between $60M to $120M for a given target. It’s probably on the lower end because not every mid-to-large biopharma company has small molecule programs. For multiple targets, this easily becomes a billion dollar opportunity.
Selling libraries of anything from small molecules to biologics to AAVs can become very unattractive if the matter within them don’t show the ability to become products. Regenxbio is in a similar market as DiCE but for AAVs - https://axial.substack.com/p/axial-regenxbio They’ve been able to garner great economics because their constructs have shown clinical success. This is similarly true in biologics licensing. So the market opportunity for selling DELs is extremely dependent on the track records of past molecules discovered using them. The results have been mixed so far with promising clinical trials but the need for approved products to validate the business of selling DELs:
Nuevolution has a molecule inhibiting BET-BD1 in the clinic - https://news.cision.com/nuevolution/r/nuevolution-s-bet-bd1-selective-inhibitor-and-candidate-compound--nue20798--shows-positive-effect-on,c2807464
HitGen has an HDAC inhibitor in the clinic for multiple myeloma - https://www.hitgen.com/enxiandao/index.php?s=/Home/Article/detail/id/448.html
GSK’s inhibitor of RIP1 (kinase) was shelved by Hal Barron - https://www.fiercebiotech.com/biotech/glaxosmithkline-s-rip1k-inhibitor-dead-arrival
X-Chem got an IND for an inhibitor of autoxin - https://www.businesswire.com/news/home/20190124005063/en/X-Rx-Announces-FDA-Acceptance-IND-Application-X-165
DELs still have some way to go.
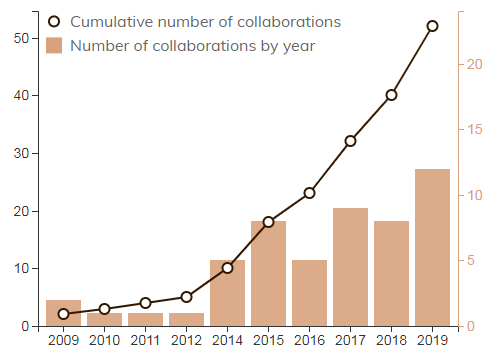
Number of DEL collaborations. Source: BPT Analytics
Business model
DiCE’s business model has evolved since its founding. A licensing model enabled a self-financing approach; however, due to the (commoditized) economics of small molecule drug development, DiCE has shifted toward an internal pipeline.
In 2013, DiCE had a really unique set of technologies in the backdrop of a DEL market that was growing pretty rapidly (the number of deals done increased 10 times from 2011 to 2015). This helped the company in 2016 to convince Sanofi to agree to a 5-year, 12-target discovery deal with $50M in up-front fees and an equity investment - https://www.reuters.com/article/us-sanofi-dice-idUSKCN0WI0KN DiCE is eligible for up to $184M in payments per target. It was an incredible deal at the time especially for a company still relatively early when compared to X-Chem and HitGen. Where HitGen was selling its DELs for up to $200K per target, DiCE got a little over $4M per target with Sanofi. That’s what you call dealing.
This set DiCE up to strike up a similar deal with Genentech in 2017 - https://www.fiercebiotech.com/biotech/genentech-dice-ally-to-go-after-hard-to-drug-targets These strategic parnterships defrayed a lot of the R&D work DiCE had to do and generated early cash flow. DiCE was positioned to scale up its licensing business, but with HitGen and so many others out there, DiCE’s DELs could become commodities. DiCE could have explored other business models like selling their mesofluidic ribosome device for a premium and commoditizing their own DELs. The company could use their DELs to develop their own drugs and license them off when they are further developed (i.e. IND, clinical-stage); they would have to add a medicinal chemistry component to their platform.
What ultimately happened was that DiCE decided to focus on an internal pipeline with their Sanofi partnership still active. DiCE can always revisit licensing out more mature assets; however, the market opportunity for DELs was on the path to commoditization compelling the company to develop its own medicines. Antibody licensing businesses have had more attractive economics on their deals because they have a history of better clinical outcomes. DELs don’t have that track record just yet. Maybe DiCE Molecules becomes DiCE Medicines.
DiCE’s lead program focuses on developing a small molecule inhibitor of interleukin-17 (IL-17) to treat psoriasis - https://www.dicemolecules.com/pipeline/il-17 There are two approved IL-17 antibodies: secukinumab (Novartis) and ixekizumab (Eli Lilly) generating over $3B and $800M in annual sales, respectively. This is a much larger market than purely selling DELs. Overall, the thesis for DiCE is to replace these biologics with a small molecule targeting IL-17. If successful, their medicine has the advantages of oral delivery: give a patient a pill instead of an IV and make it easier to combine it with other medicines.

DiCE pipeline. Source DiCE Molecules
DiCE was founded with the mission to transform small molecule discovery. They brought a new feature, directed evolution, to a validated technology, DELs. DiCE used this to strike up a unique deal that financed a lot of their internal development at very favorable, non-dilutive terms. DiCE is a case study on how a licensing model can be an efficient, both scientifically and financially, way to build out an internal pipeline. The next 7 years of DiCE will be driven by the ability of their DELs to find rare chemical matter that have better odds to become successful medicines with the goal of replacing approved biologics with a small molecule.
