The Scripps Research Institute is an institution completely focused on life sciences research and advancing medical work. Originally financed by Ellen Browning Scripps (part of a vast newspaper fortune) in 1924 as a medical center (she was inspired by the discovery of insulin to treat diabetes) and spinning off as a separate research institution in 1993, Scripps has been at the forefront of using new tools to make medical breakthroughs and discover new drugs. With a high concentration of world-class labs and an entrepreneurial culture, Scripps has spun out countless companies and driven the approval of multiple new medicines (the most recent from the Kelly and Powers Labs). Given the quality of the culture, Scripps is positioned to support important companies.

Schultz Lab
Diversity-based approaches in chemistry.Recent
Peter Schultz is a legend in chemical biology. Having worked with Christopher Walsh who worked with E.O. Wilson, it was only natural for Schultz to pioneer the combination of chemistry with evolution. Schultz has invented various methods that are taught in every chemical biology graduate course and mentored some of the field’s leaders from David Liu, Chris Anderson to Alice Ting and Kevan Shokat. Peter has been able to form a string of companies around his lab’s work from Wildcat Discovery Technologies to Ambrx and Symyx Technologies. Schultz got his PhD from Caltech in 1984 and moved to UC Berkeley where he pioneered the field of combinatorial chemistry - testing millions of molecules for drug-like properties in a high-throughput manner. He went to join Scripps in 1999 to focus more on translational research leading Genomics Institute and Calibr along the way.
Two recent papers using genomic recoding to show that unnatural amino acids are powerful to improve protein stability. GRO and Synthorx are probably the most rigorous companies using these tools for new medicines https://pubs.acs.org/doi/abs/10.1021/acschembio.9b00002 and https://pubs.acs.org/doi/abs/10.1021/jacs.8b07157
Characterizing the secretome, the set of secreted proteins using barcodes and next-generation sequencing (NGS) to identify hits - https://cell.com/cell-chemical-biology/retrieve/pii/S2451945617301800

Past
The Schultz Lab has been part of many transformative papers, many worth hanging up in your house. Engineering E. coli to incorporate and express an unnatural amino acid, p-aminophenylalanine (pAF) -https://pubs.acs.org/doi/10.1021/ja0284153 - reliant on amber suppression:

Pioneering directed evolution (David Liu took the baton forward very himself, inventing a continuous method ~10 years from this paper) - https://www.ncbi.nlm.nih.gov/pmc/articles/PMC27927/ - where enzymes now could get functionally cloned based on function:
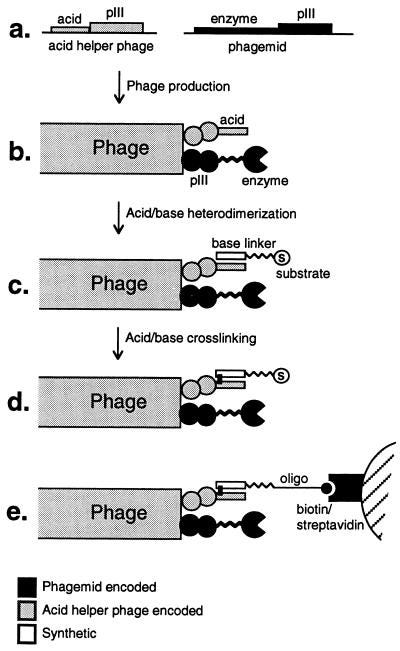
Sharpless Lab
Selectively controlling chemical reactions.Recent
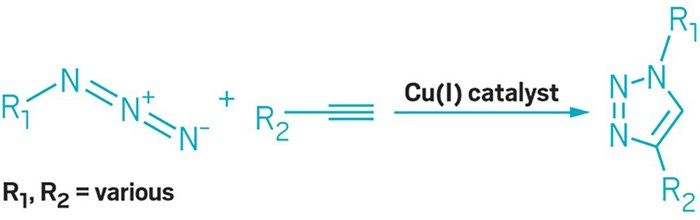
Sharpless won a Nobel for pioneering work for stereoselective chemistry that formed the basis for click chemistry. Trained at Stanford where he worked on cholesterol biosynthesis with Eugene Tamelen and organometallic chemistry with James Collman. Moving to MIT in 1969 to work with Konrad Bloch to research enzymes. In 1970, he join the MIT faculty, went to Stanford after 7 years, and came back to MIT after 3 years at Stanford. His early career focused on transforming the flat C=C bond into a 3D molecule called asymmetric epoxidation. One of his students, Eric Jacobsen (taught me organic chemistry) expanded upon this work himself to tolerate a more diverse set of functional groups. This is the work that earned Sharpless a Nobel. In 1990, he moved to Scripps and began the work that used asymmetric chemistry to invent click chemistry (who his wife actually helped coin) - a toolkit to form any bond especially in water at room temperature; incredibly useful in biological systems. The canonical example is copper-catalyzed azide-alkyne cycloaddition (CuAAC) - the reactions brings a azide and an alkyne together to form a new bond (image below). Sharpless used this reaction as the first example of click chemistry. Sharpless and his group brought together decades of research to invent a tool that abstract away a lot of complicated chemistry to allow a biochemist to link a protein to a reporter, a material scientists to link molecules to a surface, and created a whole new class of chemistry.
Inventing another biocompatible click chemistry method reliant on SuFEx transformations - https://onlinelibrary.wiley.com/doi/abs/10.1002/anie.201902489 - enabling easy S-N/S-O bond formation in cells. “Reactions that are commonly used and compatible for [DNA-encoded library] chemistry:”


Past
A broad overview of using click chemistry to easily create C-X bonds, the building block for nature and various macromolecules - https://onlinelibrary.wiley.com/doi/full/10.1002/1521-3773%2820010601%2940%3A11%3C2004%3A%3AAID-ANIE2004%3E3.0.CO%3B2-5?sid=nlm%3Apubmed

Powers Lab
Studying protein folding and aggregation.Recent
With Sharpless, describing inverse drug discovery a target-agnostic method where a electrophilic molecule is screened against a set of proteins or cell lysates. Hits are targets that bind the molecule and the drug hunter then determines if the target is valuable whereas conventional drug discovery tries to figure out if the molecule is valuable. Covalent drugs can be dangerous because they form direct bonds with their targets and could generate serious off-target effects. As a result, most approved covalent drugs have been retrospectively discovered as so - inverse drug discovery is a useful framework to rationally execute these serendipitous processes - https://pubs.acs.org/doi/abs/10.1021/jacs.7b08366 - validating this method against 11 proteins and inventing a few covalent drug hits.
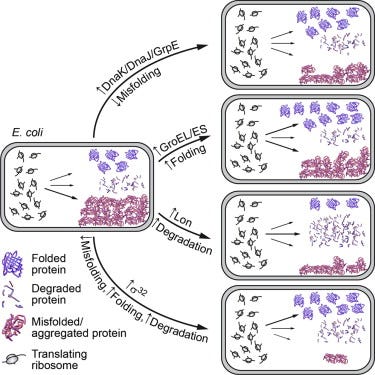
In E. coli, helping establish the set of rules for protein folding - https://www.cell.com/cell-reports/fulltext/S2211-1247(15)00271-5
Past
The paper forming the basis for the recent drug approval for cardiomyopathy - invented a screening process to discover molecules against the serum protein transthyretin (TTR) - https://stm.sciencemag.org/content/3/97/97ra81.long - where the assay measure fluorescence polarization that is amplified upon binding and minimized via tumbling along with the protein:

Law Lab
Inventing broadly protecting antibodies to ever-changing viruses.Recent
When a virus first enters the body, an immune response is mounted and memory cells are stored in preparation for the next invasion. Viruses change their out coating to subvert immune recognition making it difficult to adapt as the immune system is always anchored to the response to the first infection. This is where studying unique immune responses gets interesting - our repertoires are unique when compared to anyone. So using single-cell genomics and other tools to look into one-of-a-kind immune responses can potentially discover broad-acting antibodies with the potential to provide a long-lasting vaccine to viruses. Moreover, this work has incredible value to make new medicines for Alzheimer’s and many other diseases. Companies like Evaxion and others are doing interesting work around this problem set.
Really interesting paper to set off designing proteins that can structurally change but maintain the same function. Important to invent protein therapeutics that are non-human/humanized subverting the human immune system and enabling translation of microbial molecular tools into new medicine - https://www.biorxiv.org/content/biorxiv/early/2018/01/10/245985.full.pdf - delivery S. pyogenes and S. aureus Cas9s (chosen because of predicted lack of MHC overlap) via AAV and validating the lack of cross-reacting antibodies agains the two Cas9s in mice:

Review of vaccine design against hepatitis C - https://www.frontiersin.org/articles/10.3389/fimmu.2018.01315/full - providing a good framework mainly around structure-guided design for viruses in general (i.e. X-Vax).
Doing a study to discover broad-acting antibodies against hepatitis C- https://www.pnas.org/content/pnas/115/29/7569.full.pdf - these super-antibodies are an important approach to develop universal vaccines.
Hansen Lab
Studying the molecular basis of pain with a focus on mechanosensation, the basis of touch and sound.Recent
Showing that TREK-1, a receptor with a role in sensation that is a target of anesthetics, activation is heavily influenced by the thickness of the plasma membrane -https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3155650 - it’s still not well understood how mechanosensative receptors transmit information; this study on TREK-1 acts as a model to characterize the biophysical mechanism.
Related to the TREK-1 work, showed that anesthetics disrupt lipid rafts (another interesting biological object; Erin O’Shea taught me this section of LS1A) to influence mechanosensitivity - https://www.biorxiv.org/content/10.1101/313973v2
Lander Lab
Using cryo-EM to study molecular machines.Recent
Cryo-electron microscopy (cryo-EM) has been in a renaissance. The tool was transformed structural biology by making historically challenging molecules to study into tractable ones. From first determining the structure of the T4 bacteriophage tail to characterizing a wide array of systems, cryo-EM is being driven by advances in image-processing, more precise instruments, better electron-detection cameras, improved sample prep, and increasing automation. When compared to X-ray crystallography, the work horse for structural biologists, cryo-EM doesn’t need crystals (although crystals are still very useful), just a relatively small amount of sample. The sample will have a molecule in various conformations where cryo-EM will capture functional macromolecular complexes in different states. Whereas, X-ray crystallography captures one state. In cryo-EM, 3D to be more specific, a transmission electron microscope (TEM) takes multiple views that are reconstructed (mostly via Fourier inversion methods). This method has been very powerful for helical macromolecules that actin and phage tails. The long-term opportunity for cryo-EM is developing better methods to handle the large computational loads and using the tool to pursue hard drug targets.

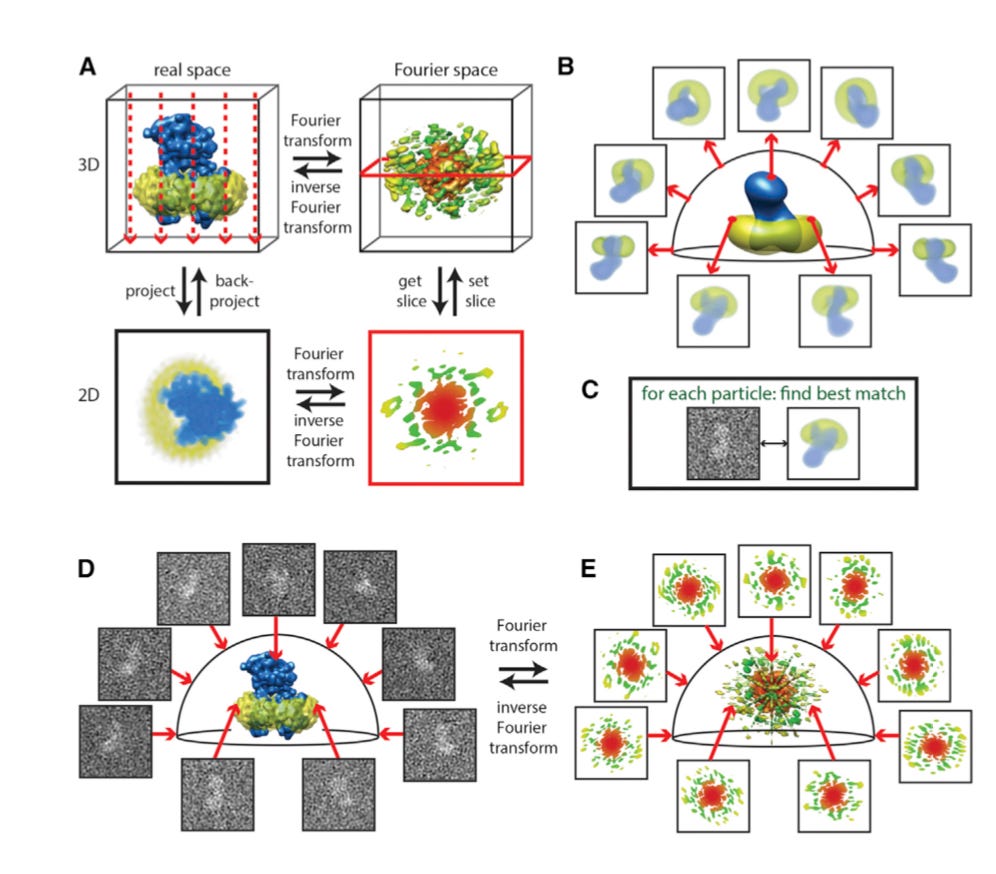
Using cryo-EM to determine the structures of smaller macromolecules (i.e. below 100 kDa) has been pretty challenging. The Lander Lab made a recent set of breakthroughs by removing the phase plate and optimizing the amount of underfocus and TEM configuration - https://www.nature.com/articles/s41467-019-08991-8 - as cryo-EM gains more value for smaller structures, its capacity to aid structure-based drug design will become more powerful.
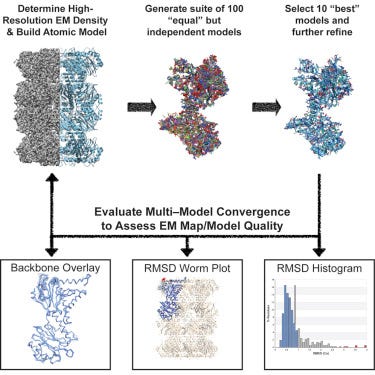
Invented a method (where there are still many bespoke tools) to accurately analyze cryo-EM data to accurately reconstruct a targets density - https://www.cell.com/structure/fulltext/S0969-2126(18)30364-2
Past
With Eva Nogales (just saw her last evening eating at a restaurant in Berkeley I was in with grad school buddy), a legend, did an overview of cryo-EM and its use to complement other biophysical techniques - https://www.ncbi.nlm.nih.gov/pubmed/22835744
Using unnatural amino acid labeling (a tool the Schultz Lab pioneered), to selectively identify protein subunits during a cryo-EM run - https://www.ncbi.nlm.nih.gov/pubmed/26409249 - allowing easier internal domain labeling where most method focus on termini:
Burton Lab
Pioneering rational vaccine design.Recent
Generating an incredible repository of data on circulating B-cells in 10 patients - https://www.nature.com/articles/s41586-019-0879-y - the outstanding problem in the field though is measuring the whole B-cell population both circulating and peripheral:

Interesting review on broad-acting antibodies (i.e. super-antibodies) - https://www.nature.com/articles/nri.2017.148