Axial: https://linktr.ee/axialxyz
Axial partners with great founders and inventors. We invest in early-stage life sciences companies such as Appia Bio, Seranova Bio, Delix Therapeutics, Simcha Therapeutics, among others often when they are no more than an idea. We are fanatical about helping the rare inventor who is compelled to build their own enduring business. If you or someone you know has a great idea or company in life sciences, Axial would be excited to get to know you and possibly invest in your vision and company. We are excited to be in business with you — email us at info@axialvc.com
This paper presents a novel approach to using large language models (LLMs) like GPT-4 for the task of matching patients to clinical trials based on their eligibility criteria. The authors demonstrate state-of-the-art performance on a benchmark dataset while highlighting the efficiency and interpretability advantages of their zero-shot LLM-based system.
The authors show that GPT-4, without any fine-tuning or in-context examples, can outperform previous state-of-the-art methods on the 2018 n2c2 cohort selection challenge benchmark. This is significant as it demonstrates the potential for LLMs to be rapidly deployed for diverse clinical trials without extensive customization.
By having clinicians evaluate the rationales generated by GPT-4 for its decisions, the authors demonstrate the potential for human-in-the-loop oversight and collaboration with LLM-based systems. The authors develop a two-stage pipeline using smaller embedding models to pre-filter relevant clinical notes before processing with the LLM, showing how this can maintain performance while significantly reducing token usage.
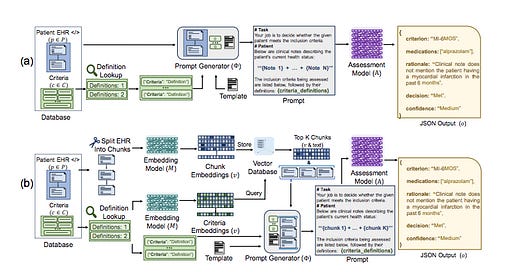
The authors evaluate several LLMs (GPT-3.5, GPT-4, Llama-2, Mixtral) on the task of determining whether patients meet specific eligibility criteria for clinical trials based on their electronic health records (EHRs). They test four main prompting strategies:
1. All Criteria, All Notes (ACAN)
2. All Criteria, Individual Notes (ACIN)
3. Individual Criteria, All Notes (ICAN)
4. Individual Criteria, Individual Notes (ICIN)
These strategies vary in how they chunk the patient notes and criteria into prompts for the LLM. The authors also develop a two-stage retrieval pipeline that uses smaller embedding models (BGE or MiniLM) to pre-filter the most relevant chunks of clinical notes before passing them to the LLM.
GPT-4 achieved state-of-the-art results on the n2c2 benchmark, surpassing the previous best model by 6 points in Macro-F1 and 2 points in Micro-F1 score. This was achieved without any fine-tuning or in-context examples, demonstrating the strong zero-shot capabilities of advanced LLMs for this task. There was a significant performance gap between GPT-4 and other models tested (GPT-3.5, Llama-2, Mixtral). This aligns with other recent studies showing GPT-4's superior capabilities on complex reasoning tasks.
The ACIN (All Criteria, Individual Notes) strategy provided the best balance of performance and efficiency. It achieved high accuracy while using significantly fewer tokens and API calls compared to strategies that evaluated criteria individually. The two-stage retrieval approach using embedding models to pre-filter relevant chunks of clinical notes showed promise in reducing token usage. With the MiniLM embedding model, they were able to surpass the prior state-of-the-art on Macro-F1 using roughly one-third to one-half as many tokens as needed in the vanilla strategies. When GPT-4 made correct eligibility decisions, clinicians judged 89% of its rationales as fully correct and 8% as partially correct. Even for incorrect decisions, 67% of rationales were judged as correct, suggesting nuanced disagreement rather than clear errors in many cases. The authors estimate their GPT-4 based system could evaluate patients at a cost of $1.55 per patient, compared to prior estimates of $34.75 per patient for manual screening in Phase III trials. The system was also an order of magnitude faster, screening all 86 test patients in about two hours versus the expected one hour per patient for manual review.
This work demonstrates the potential for LLMs to significantly accelerate and reduce costs in clinical trial patient recruitment, a critical bottleneck in drug development. The zero-shot capabilities shown here are particularly notable, as they suggest such systems could be rapidly deployed for diverse trials without extensive customization or data labeling. The interpretability aspect, with LLMs providing rationales for their decisions, offers a path towards human-AI collaboration in this domain. This could allow for more efficient workflows where AI systems handle initial screening at scale, with human experts reviewing the rationales for key decisions.
The detailed efficiency analysis provides valuable insights for practical deployment considerations. By showing how different prompting strategies and retrieval pipelines impact performance and cost, the authors offer a framework for optimizing LLM use in resource-constrained healthcare environments.
The n2c2 benchmark, while the largest public dataset for this task, is still much smaller than real-world EHR systems. This underscores the need for efficient retrieval methods that can scale to millions of patient records. And the n2c2 challenge only covers inclusion criteria, is limited to English, and may not fully represent the complexity of real-world trial matching scenarios.
While achieving state-of-the-art results, further evaluation is needed to assess the safety and potential failure modes of deploying such systems in production healthcare environments. The reliance on models like GPT-4 may limit deployment options for health systems without access to HIPAA-compliant cloud infrastructure.
Additionally, the authors suggest that LLMs could potentially be applied to improve the design of eligibility criteria themselves, addressing another key challenge in clinical trial recruitment. By demonstrating strong performance on this task, the paper adds to the growing body of evidence that LLMs can be powerful tools for processing and reasoning over complex medical information. This has potential implications far beyond trial matching, potentially impacting areas like clinical decision support, automated chart review, and personalized treatment planning.
However, the work also highlights important challenges in deploying AI systems in healthcare, particularly around efficiency, interpretability, and safety. The authors' focus on these practical considerations provides a valuable template for future work aiming to bridge the gap between AI research and clinical deployment.
This paper presents a compelling case for the use of LLMs in clinical trial patient matching, demonstrating state-of-the-art performance with a zero-shot approach that offers significant efficiency and interpretability advantages. By carefully considering prompting strategies, retrieval methods, and providing a detailed analysis of costs and token usage, the authors offer valuable insights for practical deployment of LLMs in healthcare settings.