Unbiased mapping of cereblon neosubstrate landscape by high-throughput proteomics
Inventors & their inventions
Axial: https://linktr.ee/axialxyz
Axial partners with great founders and inventors. We invest in early-stage life sciences companies such as Appia Bio, Seranova Bio, Delix Therapeutics, Simcha Therapeutics, among others often when they are no more than an idea. We are fanatical about helping the rare inventor who is compelled to build their own enduring business. If you or someone you know has a great idea or company in life sciences, Axial would be excited to get to know you and possibly invest in your vision and company. We are excited to be in business with you — email us at info@axialvc.com
This work invents a high-throughput proteomics-based platform for the unbiased discovery and validation of molecular glue degraders (MGDs) and their neosubstrates. MGDs are small molecules that hijack the ubiquitin-proteasome system to degrade target proteins, offering a novel therapeutic approach for previously undruggable targets. However, traditional MGD discovery methods, primarily relying on cell viability assays, often overlook non-essential proteins with potential therapeutic value. This study addresses this limitation by employing a comprehensive, high-throughput proteomic screening approach.
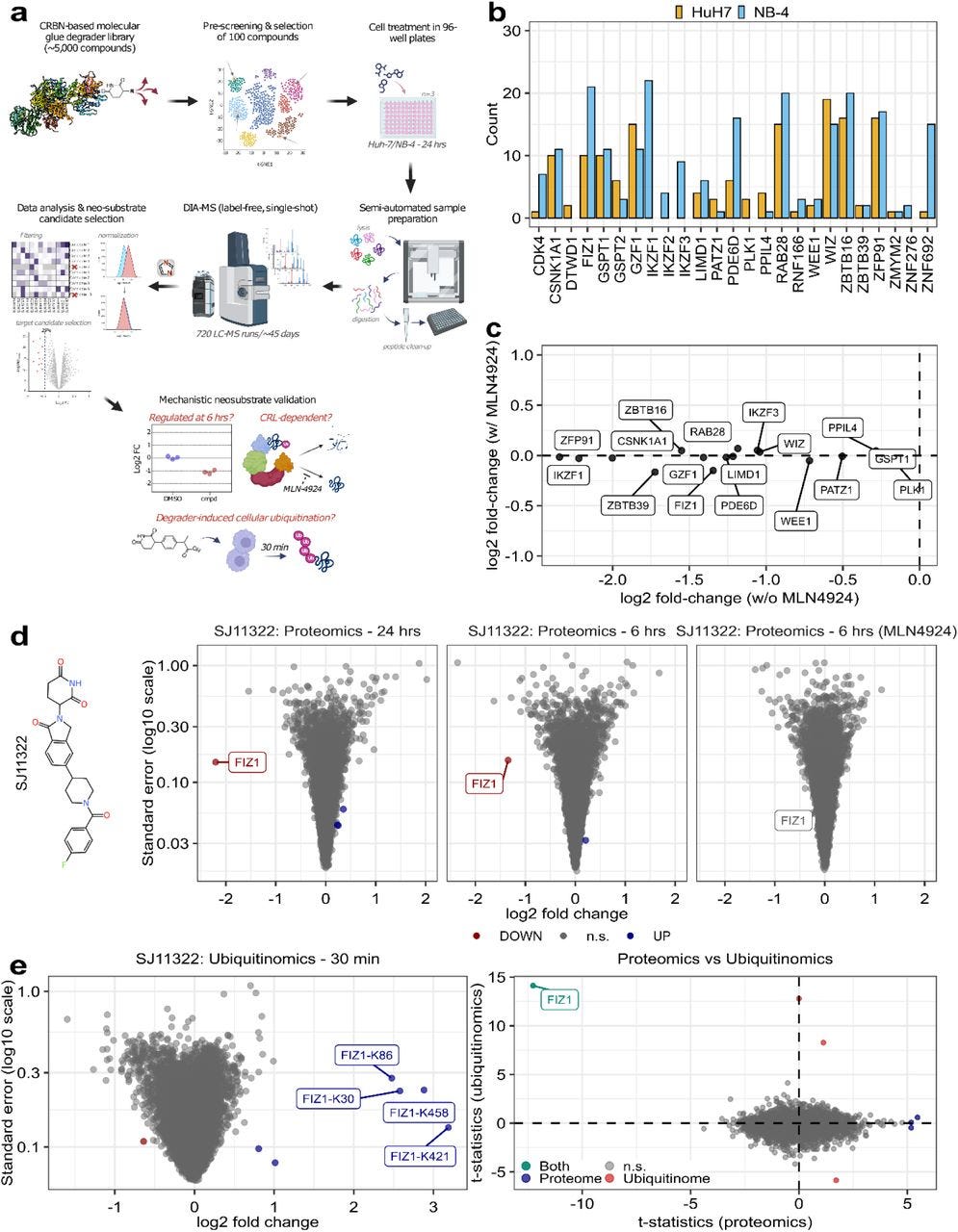
The researchers designed and synthesized a chemically diverse library of over 5,000 CRBN ligands, the majority of which possessed classical IMiD core structures (like lenalidomide and thalidomide) or novel phenyl glutarimide (PG) cores. They selected a representative subset of 100 compounds for in-depth analysis based on scaffold diversity and other chemical properties, excluding those known to target previously identified neosubstrates like GSPT1 and CK1α. The chosen compounds were tested in two cancer cell lines (Huh-7 and NB-4) known to have different protein expression profiles, maximizing the chance of identifying diverse targets.
A sophisticated multi-layered proteomics workflow was developed for high-throughput screening. This involved label-free, data-independent acquisition mass spectrometry (DIA-MS) for both proteomics and ubiquitinomics analyses. The diaPASEF method was used to maximize proteome coverage, enabling deep proteomic profiling in a highly reproducible manner. This high-throughput approach involved treatment of cells with the 100 MGDs, followed by highly sensitive MS-based proteomic and ubiquitinomics profiling. The stringent data analysis workflow focused on identifying proteins with a significant reduction (over 25%) in abundance after MGD treatment, further validated by confirming the CRL-dependency of the observed downregulation using the NEDD8 E1 inhibitor MLN4924, and finally confirmed as bona fide neosubstrates via ubiquitinomics analysis showing enhanced ubiquitination after MGD treatment.
This systematic approach revealed a remarkably broad spectrum of CRBN-mediated neosubstrates. The screen identified a substantial number of known neosubstrates, validating the platform’s efficacy and highlighting its robustness. Critically, the study also uncovered 50 novel neosubstrates, significantly expanding the known landscape of CRBN-mediated protein degradation. Notably, many of the novel neosubstrates lacked the classical CRBN degron motif, demonstrating the ability of MGDs to engage proteins beyond those previously identified based on motif recognition.
To further investigate the relationship between MGD structure and its resultant neosubstrate specificity, the researchers performed structure-activity relationship (SAR) studies. They grouped the compounds based on their core structure (IMiD-like or PG) and analyzed protein fold-changes to identify structural features associated with specific neosubstrate degradation profiles. This analysis revealed distinct selectivity patterns. For instance, IMiD-like structures predominantly targeted previously known IMiD neosubstrates such as IKZF1, ZFP91, and RAB28, while PG-based compounds showed a high selectivity for novel neosubstrates like KDM4B and FLCN. This suggests a strong structure-activity relationship that can be exploited for the rational design of MGDs with precise target selectivity.
The study focused on two particularly interesting novel neosubstrates: G3BP2 and KDM4B. G3BP2 is an RNA-binding protein involved in stress granule formation. It interacts with other proteins such as USP10, forming a complex. The researchers identified SJ41824, a potent PG-based degrader of G3BP2, displaying high selectivity. Through chemical optimization, a more potent analogue, SJ41813, was discovered which showed superior G3BP2 degradation efficacy and confirmed G3BP2 as the direct target, with USP10 showing a secondary, bystander degradation effect.
KDM4B is a histone demethylase involved in chromatin regulation, a key area of interest in cancer drug discovery, however, selective KDM4B inhibitors have been challenging to develop. The researchers found SJ41564, a PG-based MGD, selectively degraded KDM4B with high potency and selectivity. Its degradation was CRBN-dependent, and through SAR analysis, they identified several potent analogues. Interestingly, the discovery of KDM4B and G3BP2 degraders with high selectivity suggests that these less-studied proteins may prove to be crucial drug targets, particularly in the context of cancer therapy.
Furthermore, the study extended the analysis to another novel neosubstrate, VCL (Vinculin), which was found to be selectively degraded by several para-substituted PG analogues. The high selectivity of these compounds, particularly for VCL, suggests these are also promising new drug targets.
This study successfully demonstrates a high-throughput proteomic platform for the unbiased identification and validation of MGDs and their neosubstrates. The identification of several novel neosubstrates without classical degron motifs highlights the wide range of potential therapeutic targets amenable to MGD-mediated degradation. The integrated approach, combining proteomics, ubiquitinomics and SAR analyses, provides a robust framework for rational MGD design and optimization, moving drug discovery beyond traditional approaches focused primarily on druggable pockets. The discovery of highly selective and potent degraders for G3BP2, KDM4B, and VCL underscores the immense potential of this approach for tackling previously undruggable targets and advancing the field of targeted protein degradation. The extensive data generated in this study, including the comprehensive characterization of the novel MGDs and their neosubstrates, represents a substantial contribution to the development of more effective and selective therapeutics for a variety of diseases.