Single molecule imaging of the central dogma reveals myosin-2A gene expression is regulated by contextual translational buffering
Inventors & their inventions
Axial: https://linktr.ee/axialxyz
Axial partners with great founders and inventors. We invest in early-stage life sciences companies such as Appia Bio, Seranova Bio, Delix Therapeutics, Simcha Therapeutics, among others often when they are no more than an idea. We are fanatical about helping the rare inventor who is compelled to build their own enduring business. If you or someone you know has a great idea or company in life sciences, Axial would be excited to get to know you and possibly invest in your vision and company. We are excited to be in business with you — email us at info@axialvc.com
This paper by O’Neil Wiggan and Timothy Stasevich details the creation and application of a novel human cell line (eMyo2AGFP) to study the regulation of the MYH9 gene, which encodes myosin-2A, with unprecedented single-molecule precision. The authors achieve this by employing CRISPR-Cas9 gene editing to introduce multiple fluorescent tags into the endogenous MYH9 locus, allowing simultaneous visualization of transcription, translation, mRNA, and mature protein. This innovative approach provides a powerful tool to dissect the central dogma in living cells, moving beyond the limitations of traditional methods.
The introduction effectively establishes the significance of understanding gene expression homeostasis, particularly focusing on the challenges in determining regulatory mechanisms at different levels of the central dogma. The authors highlight the ambiguity surrounding MYH9 regulation, given its dual categorization as both a housekeeping gene (suggesting stable expression) and a gene implicated in cancer (suggesting dynamic regulation). This sets the stage for their innovative approach, emphasizing the need for a system capable of directly imaging all aspects of gene expression with single-molecule resolution.
The results section systematically demonstrates the capabilities of the eMyo2AGFP cell line. The authors meticulously validate the functionality of their tagged MYH9 gene, confirming its proper localization and expression levels comparable to untagged controls. They then demonstrate the ability to resolve individual transcription events, quantify mRNA numbers and localization, and precisely identify and characterize translation sites using both fixed-cell immunostaining and live-cell imaging with anti-FLAG intrabodies. This detailed characterization establishes the cell line's robustness and sensitivity for studying MYH9 expression dynamics.
The study proceeds to investigate MYH9 expression regulation across different cellular contexts. Analysis of MYH9 expression throughout the cell cycle reveals a dynamic relationship between transcription and translation, with a marked decrease in translation efficiency during mitosis, independent of mRNA levels. This observation suggests a cell cycle-dependent regulatory mechanism fine-tuning myosin-2A expression to avoid potential disruptions to cell division. The authors carefully consider and rule out alternative explanations, like changes in cell size, supporting the conclusion of cell cycle-dependent translational regulation.
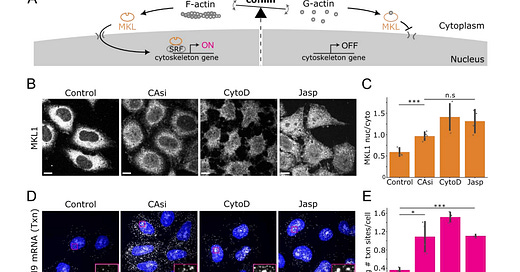
Further exploration of MYH9 regulation focuses on the impact of actin depolymerizing protein knockdown. The authors observe a dramatic increase in MYH9 transcription upon silencing cofilin and ADF, driven by increased transcription burst frequency and amplitude. They demonstrate that this transcriptional upregulation is largely dependent on Serum Response Factor (SRF), a key regulator of cytoskeletal gene expression. This finding establishes a direct link between disrupted actin dynamics, increased MYH9 transcription, and the potential for detrimental cellular consequences.
Interestingly, the authors uncover a mechanism of translational buffering in response to this transcriptional upregulation. While cofilin/ADF knockdown leads to a significant increase in MYH9 mRNA, the increase in myosin-2A protein is considerably less pronounced. They demonstrate that this buffering effect is achieved by a reduction in the fraction of cytoplasmic mRNA undergoing translation, rather than changes in individual translation site intensity. This suggests a mechanism switching translation on or off for individual mRNA molecules rather than gradually modulating translation rate. The authors rule out this buffering as a general effect, confirming the specificity of the MYH9 regulation.
In contrast to the translational buffering observed with cofilin/ADF knockdown, the response to serum stimulation shows a different regulatory pattern. Upon serum stimulation, MYH9 transcription and translation increase in tandem, enabling rapid actomyosin cytoskeletal reorganization. The authors provide evidence that both SRF-dependent transcription and new protein synthesis are essential for the formation of stress fibers, but not for initial peripheral actomyosin bundling in response to serum, This finding underscores the context-dependent nature of MYH9 regulation, highlighting the adaptability of the system in response to different cellular stimuli.
The discussion section synthesizes the key findings, emphasizing the multi-layered regulatory network governing MYH9 expression. The authors effectively portray MYH9 translational regulation as a crucial homeostatic mechanism, analogous to a thermostat, ensuring appropriate myosin-2A levels. They propose a model where translational buffering acts as a safeguard against excessive myosin activity, preventing detrimental effects. The contrast between the responses to cofilin/ADF knockdown and serum stimulation highlights the context-dependent nature of this regulatory mechanism.
The authors acknowledge limitations of their approach, such as the challenge in detecting dimmer translation sites and the incomplete tagging of MYH9 alleles. They also discuss the potential for future research, including the identification of specific molecular players involved in translational buffering and further exploration of the generalizability of their approach to other cell types and genes. Despite these limitations, they convincingly argue that the eMyo2AGFP cell line represents a significant advance, offering a valuable tool for investigating the complexities of gene regulation. The concluding paragraphs effectively emphasize the broad applicability of this innovative system, highlighting its potential for future studies in diverse cellular contexts. Overall, the paper presents a well-executed and impactful study that significantly advances our understanding of gene expression regulation.