Proteomics
Call for talented individuals and teams
Axial: https://linktr.ee/axialxyz
Axial partners with great founders and inventors. We invest in early-stage life sciences companies such as Appia Bio, Seranova Bio, Delix Therapeutics, Simcha Therapeutics, among others often when they are no more than an idea. We are fanatical about helping the rare inventor who is compelled to build their own enduring business. If you or someone you know has a great idea or company in life sciences, Axial would be excited to get to know you and possibly invest in your vision and company. We are excited to be in business with you — email us at info@axialvc.com
Proteomics is the large-scale study of proteins, particularly their structures and functions. The term was coined in the 1990s to analogize with genomics, the study of the genome. Proteomics and genomics are intertwined sciences, as proteins are translated from messenger RNA which is transcribed from genomic DNA. However, while the genome is more or less static, the proteome differs from cell to cell and constantly changes through its biochemical interactions with the genome and the environment.
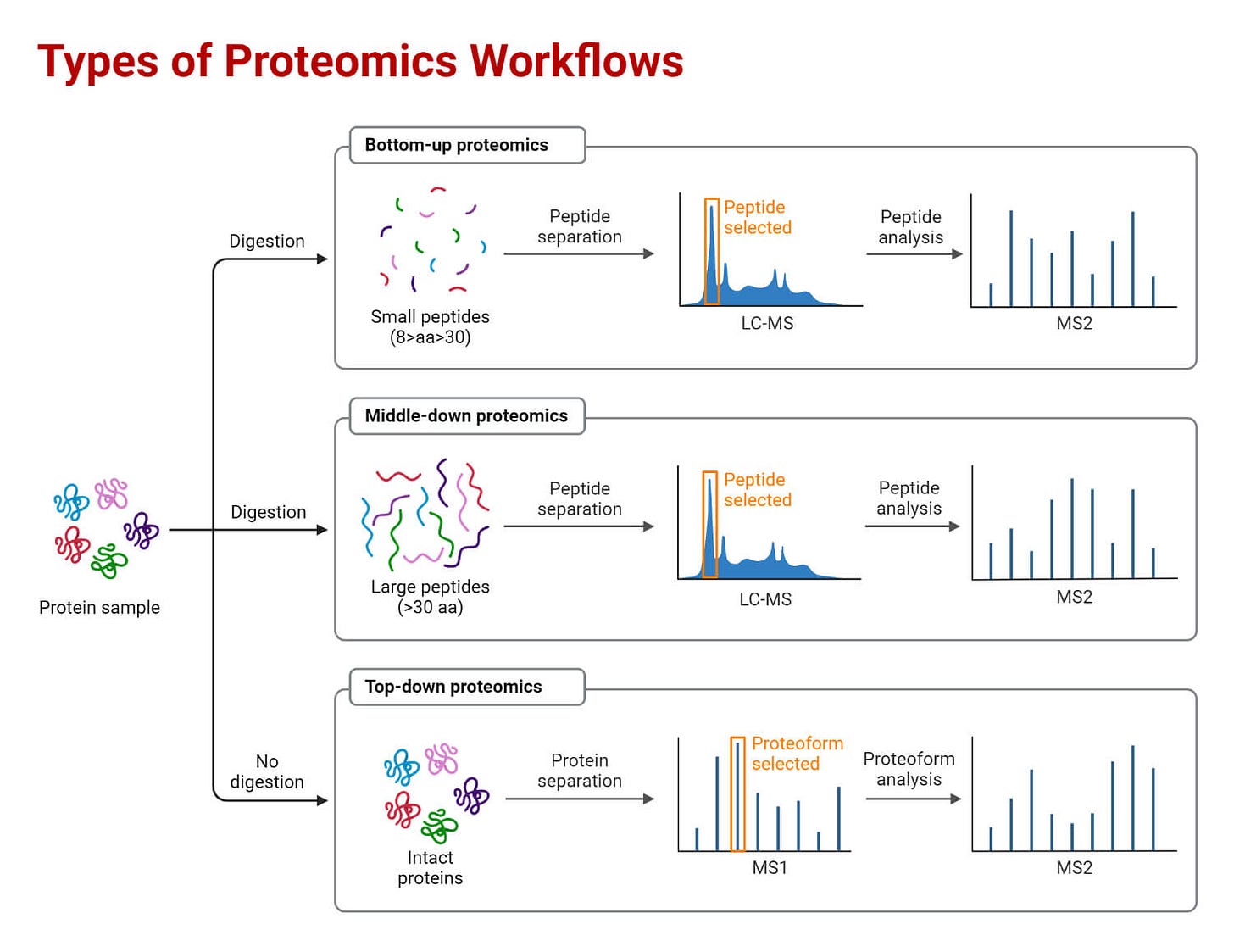
Proteomics involves the identification and quantification of proteins in biological systems. It relies heavily on mass spectrometry and bioinformatics to identify and characterize proteins and their modifications. The field has progressed tremendously in the past couple of decades, with advances in instrumentation, computational methods, and techniques allowing deeper coverage and analysis of proteomes. However, significant challenges remain in sensitivity, throughput, quantification, and data analysis.
Instrumentation: The development of soft ionization techniques like electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI) have been critical for enabling mass spectrometric analysis of proteins and proteomes. Improved mass analyzers like Orbitrap and advanced hybrid instruments that combine multiple analyzers have also boosted sensitivity and throughput.
Fragmentation techniques: Tandem mass spectrometry using collision-induced dissociation (CID), electron-transfer dissociation (ETD), and other methods provide sequence information and PTM localization. Newer techniques like ultraviolet photodissociation (UVPD) increase sequence coverage.
Separation and fractionation: Multidimensional fractionation strategies including liquid chromatography, capillary electrophoresis, and gel electrophoresis reduce sample complexity and improve proteome coverage. Reversed-phase and ion exchange chromatography are commonly used for pre-fractionation.
Quantification: Label-based (SILAC, TMT) and label-free quantification methods allow relative protein abundance to be determined across samples. This enables identification of biomarkers and changes in signaling pathways.
Bioinformatics: Faster database searching algorithms, statistical models, and machine learning approaches allow high-throughput data analysis and information extraction from large proteomic datasets. Resources like ProteomeXchange facilitate data sharing.
Single-cell analysis: Recent advances are making single-cell resolution proteomics possible through techniques like microfluidics and barcode multiplexing. This provides finer insight into cell heterogeneity.
Some key challenges holding back progress in proteomics are:
- Sensitivity - Low ionization/detection efficiency of intact proteins and limitations in MS dynamic range restrict depth of coverage. Prefractionation helps but reduces throughput.
- Throughput - Analysis of intact proteins is slower than peptides. Data acquisition rates need to improve drastically.
- Quantification - Precise quantification of large numbers of proteins across multiple samples remains difficult.
- PTM analysis - Localization of combinatorial PTMs on proteins is still challenging and requires new fragmentation techniques.
- Data analysis - Handling and making sense of massive, multidimensional proteomic datasets requires better bioinformatics tools.
- Single-cell analysis - Isolating and analyzing proteins from single cells is still technically difficult and labor-intensive.
The prospects for proteomics are bright, with various new technologies and approaches on the horizon. Novel mass analyzers like trapped ion mobility spectrometry (TIMS) promise to improve sensitivity and separations. Integrated lab-on-a-chip devices could enable automated sample processing and boost throughput. Top-down and middle-down proteomics will become more mainstream with improved fragmentation techniques. Multiplexing through isobaric tagging will enhance quantification of large proteome samples. Machine learning and data analytics will extract deeper insights from proteomic data. Single-cell techniques will become higher throughput and more quantitative. Spatial proteomics will provide context. Clinical proteomics will identify proteomic biomarkers for precision diagnostics and medicine.
However, realizing the full potential of proteomics will require a coordinated effort across disciplines and substantial investment in infrastructure and training. Data standards and sharing protocols also need to be established. Integrating proteomics with genomics and other omics data will provide a comprehensive systems-level understanding of biological organisms. Overall, proteomics remains an exciting and rapidly developing field that promises to transform biological research and medical applications over the coming decades.
Proteomics has seen remarkable progress over the past couple of decades, driven by advances in mass spectrometry, separation techniques, quantification, and computation. However, significant challenges remain in sensitivity, throughput, quantification, and data analysis. The field is poised for growth with the emergence of new technologies and approaches that will help address these challenges. Realizing the immense promise of proteomics will require interdisciplinary collaboration and major investments in infrastructure and personnel training. With dedicated efforts to overcome existing limitations, proteomics can usher in a new era of systems biology and personalized medicine in the 21st century.