Nicole Martinez, RNA Modifications & Rewriting the Message in Health & Disease
Top builders in biotech
Axial: https://linktr.ee/axialxyz
Axial partners with great founders and inventors. We invest in early-stage life sciences companies such as Appia Bio, Seranova Bio, Delix Therapeutics, Simcha Therapeutics, among others often when they are no more than an idea. We are fanatical about helping the rare inventor who is compelled to build their own enduring business. If you or someone you know has a great idea or company in life sciences, Axial would be excited to get to know you and possibly invest in your vision and company . We are excited to be in business with you — email us at info@axialvc.com
Nicole Martinez is an Assistant Professor in the Departments of Chemical and Systems Biology and of Developmental Biology at Stanford. Where her lab studies RNA regulatory mechanisms that control gene expression. Focusing on mRNA processing, RNA modifications and their roles in development and disease.
She earned her PhD studying alternative splicing with Kristen Lynch at the University of Pennsylvania, and then went on to do her postdoc with Wendy Gilbert at Yale. Where she investigated pre-mRNA pseudouridylation, and established the role of pseudouridine synthases in mRNA processing. Then opening up her own lab to study everything from splicing to 3’ end processing & RNA modifications.
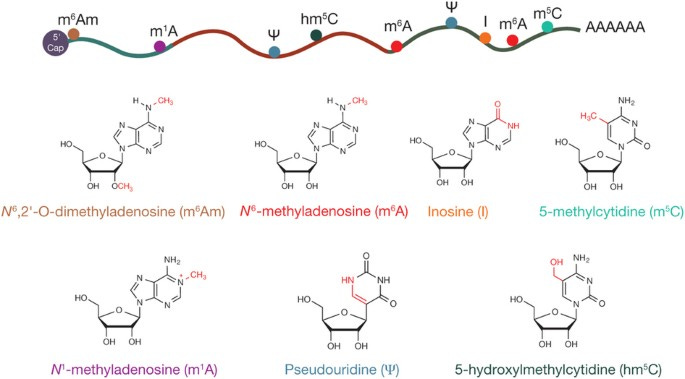
RNA modifications have been historically considered exclusive to infrastructural RNAs, such as rRNAs, tRNAs, and snoRNAs. Irreversible decorations important for RNA structural stability and/or catalytic function. With new transcriptomic tools & systematic efforts to map RNA modifications, they are found to be far more widespread than originally thought. Reversible, dynamically regulated, and involved in key biological processes. For example, N6-methyladenosine (m6A) modification has been identified as an important factor in the determination of mammalian cell fate transition and embryonic stem cell differentiation. With new findings for m1A, m6Am, m5C and pseudouridine. Creating a new field of epitranscriptomics.
The functional and evolutionary relevance of the epitranscriptome is yet unknown, but it may represent the crossroads of gene–environment interactions for physiological adaptation and cognition. There is a progressive expansion of enzymes that impart RNA editing and the extent of RNA editing in the brain during cognitive evolution. A clear example of this expansion is the family of adenosine deaminases acting on RNA (ADAR), responsible for the editing of adenosine to inosine, thus increasing gene product diversity, particularly in primates
The field is now producing detailed maps of the location and abundance of a handful of RNA modifications. Mostly from chemical treatments to next-generation sequencing or coupling antibody immunoprecipitation. Moving toward characterization of their biological function.
RNA modifications have been historically considered to be relatively static fine-tuners of the RNA structure and function. However, in the last few years, it has become evident that the epitranscriptomic layer is not only dynamic and reversible - catalyzed by RNA modification “erasers” - but regulated by a wide variety of factors, including environmental conditions.
The dysregulation of RNA modification and mRNA processing can have a profound impact on human health:
In Fragile X syndrome, a mutation in the FMR1 gene leads to a decrease in the production of the FMRP protein. FMRP is a RNA binding protein that plays a role in regulating the expression of a number of genes involved in brain development. The loss of FMRP leads to intellectual disability, developmental delays, and behavioral problems.
In Huntington's disease (HD), a mutation in the HTT gene leads to an expansion of CAG repeats in the HTT protein. The expanded HTT protein is toxic to neurons and causes their death. The death of neurons in the brain leads to the characteristic symptoms of Huntington's disease, such as chorea (involuntary movements) & cognitive decline.
In cancer, mutations in RNA modification enzymes can lead to changes in gene expression that promote tumor growth and metastasis. For example, mutations in the ALK gene can lead to the production of an abnormal ALK protein that is able to activate a number of growth-promoting signaling pathways. Leading to the development of aggressive lung cancer.