Harnessing eukaryotic retroelement proteins for transgene insertion into human safe-harbor loci
Inventors & their inventions
Axial: https://linktr.ee/axialxyz
Axial partners with great founders and inventors. We invest in early-stage life sciences companies such as Appia Bio, Seranova Bio, Delix Therapeutics, Simcha Therapeutics, among others often when they are no more than an idea. We are fanatical about helping the rare inventor who is compelled to build their own enduring business. If you or someone you know has a great idea or company in life sciences, Axial would be excited to get to know you and possibly invest in your vision and company. We are excited to be in business with you — email us at info@axialvc.com
This work by introduces a novel method called PRINT (Precise RNA-mediated Insertion of Transgenes) for inserting genes into specific locations in the human genome. The technique utilizes proteins from R2 retroelements, which are mobile genetic elements found in many animals, to insert DNA copies of RNA templates into a specific location in ribosomal DNA genes.
Key Components and Mechanism of PRINT:
1. Two RNA components are delivered into cells:
- mRNA encoding the R2 protein
- Template RNA containing the transgene to be inserted
2. The R2 protein recognizes a specific DNA sequence in ribosomal DNA genes, creates a nick in one strand, and uses that nick to prime reverse transcription of the template RNA directly into the genome.
3. It targets multicopy ribosomal DNA genes as a "safe harbor" location that can tolerate insertions without disrupting essential genes.
4. The method does not rely on DNA double-strand breaks or DNA donor templates, potentially reducing off-target effects and immune responses compared to some other gene insertion methods.
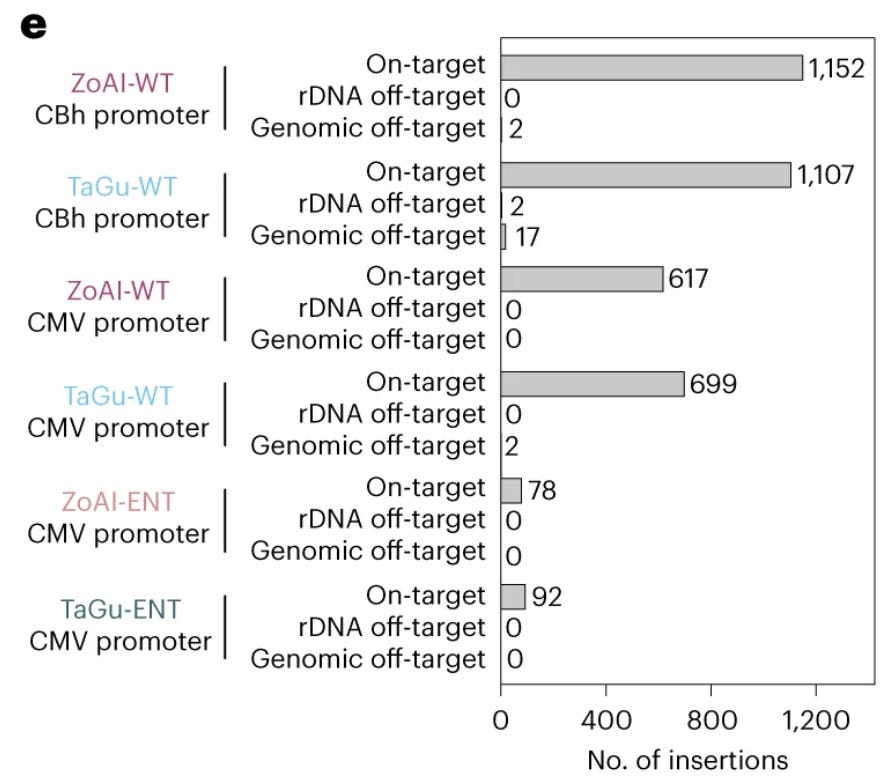
Under optimized conditions, transgene insertion was achieved in over 50% of cells, with more than 50% of insertions being full-length copies of a 2 kb transgene. The vast majority of insertions (>99%) occurred at the intended ribosomal DNA target site. Off-target insertions were rare, especially for the ZoAl R2 protein derived from the white-throated sparrow. When insertion copy number was limited (using an "ENT" version of the R2 protein with reduced endonuclease activity), transgene expression remained stable over 2 months of cell culture. The system worked in multiple human and animal cell types and could insert transgenes up to at least 4 kb in length. The use of RNA rather than DNA templates may reduce risks of random genomic insertion. The ribosomal DNA target site is considered a "safe harbor" location unlikely to disrupt other genes.
Key discoveries:
1. Identification of avian R2 proteins (especially from zebrafinch and white-throated sparrow) as having high specificity for their native RNA templates, unlike previously studied R2 proteins.
2. Discovery that these R2 proteins require a short (4 nucleotide) ribosomal RNA sequence at the 3' end of the template RNA for efficient insertion, providing an additional layer of specificity.
3. Development of a two-RNA delivery system separating the protein-coding and template functions, overcoming previous limitations of trying to use R2 elements in trans.
4. Demonstration that using RNA rather than DNA templates can lead to efficient transgene insertion while potentially improving safety.
5. Creation of "ENT" (endonuclease-tuned) versions of R2 proteins that reduce insertion frequency but improve long-term stability of transgene expression.
The authors screened multiple R2 proteins from different species for their ability to perform target-primed reverse transcription (TPRT) in vitro. They found that avian R2 proteins, particularly those from Taeniopygia guttata (zebrafinch, TaGu) and Zonotrichia albicollis (white-throated sparrow, ZoAl), showed high specificity for their native RNA templates and precise target site recognition.
The researchers discovered that efficient insertion required the template RNA to have a short (4 nucleotide) ribosomal RNA sequence at its 3' end, followed by a poly-A tail. This sequence allows base-pairing with the DNA at the target site, enhancing specificity and efficiency. By separating the R2 protein-coding sequence and the transgene template into two different RNA molecules, the authors overcame previous limitations in using R2 elements for gene insertion. This approach allows for more flexible design of the template RNA and potentially reduces safety concerns.
Using optimized conditions, the PRINT system achieved transgene insertion in over 50% of cells, with more than half of these insertions being full-length copies of a 2 kb transgene. The vast majority (>99%) of insertions occurred at the intended ribosomal DNA target site, with very few off-target events detected. Initial experiments showed that high copy number insertions led to decreased transgene expression over time. To address this, the authors created "ENT" (endonuclease-tuned) versions of the R2 proteins with reduced endonuclease activity. These ENT proteins resulted in fewer insertions per cell (typically 1-7 copies) but led to stable transgene expression over at least 2 months of cell culture.
The PRINT system was shown to work in multiple human and animal cell types, including both primary and transformed cell lines. The authors also demonstrated insertion of transgenes up to 4 kb in length, including a functional copy of the human telomerase reverse transcriptase (TERT) gene.
Detailed analysis of insertion junctions revealed insights into the mechanism of insertion:
1. The 3' end of the transgene typically joined seamlessly to the target site, guided by the short ribosomal RNA sequence at the end of the template.
2. The 5' junction showed more variability, with some insertions having precise joins, some showing duplications of target site sequence, and some exhibiting "snap-back" structures where the cDNA had primed off itself.
3. A small percentage of insertions incorporated sequences from endogenous non-coding RNAs (primarily U6 RNA), possibly due to template switching during reverse transcription.
Several aspects of the PRINT system potentially enhance its safety compared to some other gene insertion methods:
1. Use of RNA templates instead of DNA may reduce risks of random genomic insertion and lessen innate immune responses.
2. Targeting multicopy ribosomal DNA provides a "safe harbor" that can tolerate insertions without disrupting essential genes.
3. The high specificity of the R2 proteins for their target site minimizes off-target insertions.
4. The system does not rely on DNA double-strand breaks, which can lead to unwanted genomic rearrangements.
The development of PRINT represents a creative application of basic research on retroelements to address an important challenge in biotechnology and gene therapy. By repurposing the highly evolved target-site specificity of R2 elements, the authors have created a system that combines efficient gene insertion with potentially improved safety compared to existing methods.
The paper provides a comprehensive characterization of the PRINT system, from the initial selection and optimization of R2 proteins to detailed analysis of insertion outcomes and long-term stability. This thorough approach provides a strong foundation for future development and optimization of the technology.
While significant work remains to be done before PRINT could be used therapeutically, including long-term in vivo studies and development of efficient delivery methods, the system shows great promise. If its potential advantages in efficiency and safety are borne out in further studies, PRINT could become an important new tool in the gene therapy arsenal, potentially enabling new approaches to treating genetic diseases and engineering cells for therapeutic purposes.
The work also advances our basic understanding of R2 retroelement biology, providing new insights into the mechanisms these elements use to propagate themselves in host genomes. This underscores the value of basic research in driving biotechnological innovations.