Axial - University of Washington #1
Analysis of exciting University of Washington life sciences inventors and their inventions
University of Washington houses some of the world’s greatest computational biologists. Coupled with rigorous molecular biologist and clinicians, the university is an interesting space for unique businesses combining the powers of biology and chemistry with software.
Baker Lab
Linking protein structure to function.Recent
Great overview of de novoprotein design - https://www.bakerlab.org/wp-content/uploads/2019/04/Baker-2019-Protein_Science.pdf
Designed an interleukin (IL-2) to elicit a specific immune response where the cytokine was unique at each amino acid side and could be put in a boiling pot of water and still not denature - https://www.bakerlab.org/wp-content/uploads/2019/01/Silva2018_IL2-15.pdf- forming the basis of Neoleukin.
Using evolutionary dynamics to map out protein-protein interaction (PPI) maps - https://www.bakerlab.org/wp-content/uploads/2019/07/2019_Cong_ProteomeCoevolution.pdf- whereas conventional methods rely on some both of two-hybrid screening or mass spectrometry.
Past
The potential of de novoprotein design - https://www.bakerlab.org/wp-content/uploads/2016/09/HuangBoyken_DeNovoDesign_Nature2016.pdf

From metagenomic data, predicting protein structures of currently uncharacterized classes - https://science.sciencemag.org/content/355/6322/294
Relying on high-throughput gene synthesis and computational design to study protein folding across 1000s - https://www.bakerlab.org/wp-content/uploads/2017/12/Science_Rocklin_etal_2017.pdf
Using computational methods to improve enzyme design - https://www.bakerlab.org/wp-content/uploads/2019/02/Lau2018.pdf- improving the activity of a small ubiquitin-like modifier (SUMO) protease; valuable for engineering degradation into biological systems.
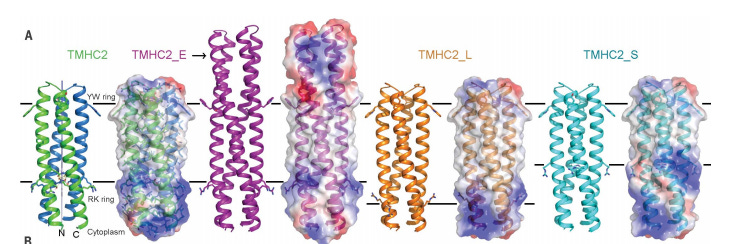
Transmembrane proteins have notoriously been difficult to crystallize due to the lipid bilayer interacting with protein segments - https://www.bakerlab.org/wp-content/uploads/2018/03/Lu_Science_2018.pdf- the Baker lab did pioneering work to not only predict transmembrane protein structures but design them:
Klavins Lab
Genetic circuit engineering.Recent
Engineering a bistable switch in yeast - https://pubs.acs.org/doi/10.1021/acssynbio.8b00524
Past
Perspective on encoding load drivers in genetic circuits to reduce cross-talk between circuits within a cell, a notorious problem limiting the potential applications of synthetic biology - http://www.nature.com/nbt/journal/v32/n12/full/nbt.3089.html

Powerful platform to use yeast mating to functionalize antibodies - https://www.pnas.org/content/114/46/12166.full- driving A-Alpha Bio:

Engineering altruism into cellular populations - https://www.biorxiv.org/content/10.1101/086900v1
Engineering finite state machinesinto genetic circuits - https://pubs.acs.org/doi/abs/10.1021/sb4001799

Chiu Lab
Developing new single-cell and single-molecule tools.Recent
Invented an integrated device to act on single-cells - https://onlinelibrary.wiley.com/doi/abs/10.1002/anie.201807314- useful for diagnostics, but more valuable for therapy sorting.
Past
Using polymer dots for long-term tracking of cancer cells in a diverse population - https://pubs.acs.org/doi/abs/10.1021/acs.analchem.7b01214- with a lot of value to track distinguishing biophysical features of cells.
Shendure Lab
Making sequencing a better tool.Recent
With Wally Gilbert himself, overview on the history of sequencing - https://shendure-web.gs.washington.edu/documents/shendure_nature_2018.pdf- this expertise is a major reason why Guardant Health bought Bellwether Bio.

Mapping out single-cell chromatin accessbility - https://shendure-web.gs.washington.edu/documents/cusanovich_cell_2018.pdf- in mice.
Past
Perspective on studying genetic variants at scale - https://shendure-web.gs.washington.edu/documents/fields_genetics_2016.pdf- driving companies like Empiricoand Variant.
Inventing a single-cell, multiplex Hi-C method to study the diversity of the 3D Genome - https://shendure-web.gs.washington.edu/documents/ramani_nature_methods_2017.pdf
Stamatoyannopoulos Lab
Understanding how genetic regulation drives disease.Past
The StamLab does pioneering work to understand the genome, in particular the 3D Genome, and how it affects disease. Now doing most of their work through Altiusin partnership with GSK to develop drugs altering the 3D Genome. Led the ENCODE project to map out the regulatory elements found in the genome - https://www.nature.com/articles/nature11247
Mapping out the chromatin landscape of the genome - https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3721348/
Overview of tools measuring single-cell genetic regulatory variation - https://www.cell.com/cell-systems/fulltext/S2405-4712(15)00019-8
Inventing a unique method to piggy back on gene editing tools to map out functional in the regulatory regions of the genome -https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5381659/- at a nucleotide resolution.
Large-scale study of in vivodynamics between gene variants and their corresponding transcription factors - https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4666772/
Wiggins Lab
Single-cell studies to understand biological structure and function.Recent
Using the bacterial cytoplasm to do profound work to understand scale invariance of biological phenomenba - https://journals.aps.org/pre/abstract/10.1103/PhysRevE.97.062410
Past
Developing an automated fluorescence microscopy method - https://onlinelibrary.wiley.com/doi/full/10.1111/mmi.13591- that can measure an order of magnitude for cell-features. Useful as sorting shifts from extracellular markers to intracellular ones (10% versus 100% of addressable space).
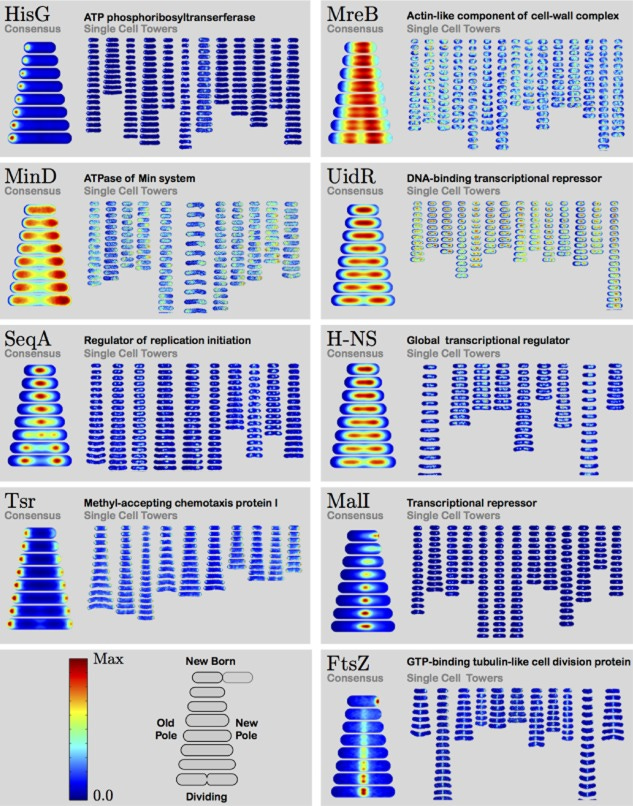
Doing outstanding work to map out the temporal and spatial localazation of each protein - https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4309519/- across the cell cycle in E. coli; calling the data set the localizome:
Studholme Lab
Computational analysis to study brain anatomy.Recent
In utero, mapping out fetal white matter brain connectivity - https://www.spiedigitallibrary.org/conference-proceedings-of-spie/10578/1057809/Comparing-diffusion-tensor-and-spherical-harmonic-tractography-for-in-utero/10.1117/12.2294476.short?SSO=1
Past
Creating a model to integrate multiple MRI views to create a reconstruction algorithm that does not have to be anchored to a prior - https://www.sciencedirect.com/science/article/abs/pii/S1361841513001552
Overview of modeling the human fetal brain - https://www.sciencedirect.com/science/article/abs/pii/S0736574813001032

Kueh Lab
Studying immune cell decision making.Recent
Determined that the states of T-cells are determined by long-range parts of the genome (i.e. 3D Genome) - https://elifesciences.org/articles/37851
Past
Reconstructed the T-cell lineage regulatory network - https://onlinelibrary.wiley.com/doi/full/10.1002/wsbm.162- setting the base to engineer T-cell fate decision making:




