The Salk Institute probably has the best lab space in the world - designed by Louis Kahn. On the coast of La Jolla, great science is done in spite of the awesome surfing and sunny days. Founded by Jonas Salk, the inventor of the polio vaccine, the institute perennially produces world-class discoveries and inventions.

Hsu Lab
Inventing tools to edit biology.Recent
The Hsu Lab is a pioneer in gene editing - despite Patrick Hsu recently joining the bioengineering faculty at UC Berkeley, it’s still appropriate to include him with the Salk since his most recent work came out of the institution. The future is bright for the lab and it’s almost a guarantee that their work at UC Berkeley will transform gene editing and the world. Recently, the lab developed a high-throughput pairwise screening method to investigate SaCas9 activity and their corresponding guides (screened over 80K different guides) -https://www.nature.com/articles/s41467-018-05391-2

Using metagenomics, nature is an incredible tinkerer/inventor, the Hsu Lab discovered a new class of CRISPR with an effector named CasRx with a size of 930 amino acids and the ability to edit RNAs - https://hsu.salk.edu/papers/Konermann_Cell_2018.pdf - showed major advantages in AAV delivery with a proof-of-concept modulating splicing for a Tau isoform. Can easily form the basis of a new company with others like Shape and Scribe working on similar stuff:


Past
One of the most important papers in gene editing - showing the use of Streptococcus pyogenes Cas9 (SpCas9) to edit genes in human cells - truly incredible work - https://hsu.salk.edu/papers/Hsu_PD_Nat_Biotechnol_2013.pdf
Saghatelian Lab
Studying the role of natural small molecules in physiology and disease.Recent
Alan Saghatelian is one of the best chemical biologists in the world. Trained at UCLA and Scripps, his work has showed the power of bringing chemical tools to study biology. Now at the Salk, Harvard Chemistry made a horrible mistake but that’s a personal opinion and an aside - Alan taught me chemical biology, the Saghatelian Lab is positioned to discover entirely new classes of natural small molecules and their role in disease. New medicines and great companies will surely get formed over time. Recently, the lab discovered the role of a 54 amino acid microprotein, PIGBOS, in the unfolded protein response (UPR). Personally, what’s pretty awesome is seeing the initial kernel of ideas explored many years ago actually lead to a foundational discovery -https://www.nature.com/articles/s41467-019-12816-z

Past
Collaborating with the Lee Lab, make a really interesting discovering for a new mechanism-of-action driving Alzheimer’s. Apolipoprotein E (ApoE) is a gene discovered through GWAS studies to be protective against the disease in particular the ε4 (E4) variant - https://pubs.acs.org/doi/10.1021/jacs.6b03463 - based on previous studies that showed a serine protease processed ApoE4 and Tau and amyloid precursor proteins are targets of a serine protease named high-temperature requirement serine peptidase A1 (HtrA1), the Saghatelian Lab hypothesized that HtrA1 may act on ApoE4. Based on that hypothesis, they figured out that HtrA1 does selectively act on ApoE4 and actually prevent that protease from acting on Tau, acting as a sink, providing a new target to treat Alzheimer’s.
Hargreaves Lab
Researching the role of chromatin remodeling in disease.Recent
BRG1-associated factors (BAF), a chromatin remodeler, has a major influence on stem cell proliferation - the Hargreaves Lab discovered that two proteins, BRD9 and GLTSCR1, form a non-canonical (i.e. not normally observed) BAF complex to drive pluripotency - https://www.nature.com/articles/s41467-018-07528-9 - the quality of work to characterize this is outstanding and pushes the regenerative medicine field forward toward a deeper understanding of stem cells.
Past
Using ATAC-seq to explore the chromatin landscape in colorectal cancer cells with and without ARID1A/B proteins, both having roles in cancer, to discover that ARID1A drives overall accessibility of enhancers - https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5643100/
Navlakha Lab
Biology-inspired algorithms.Recent
The Navlakha Lab is doing really cool research to use biology to invent new algorithms; I have a buddy at DeepMind who is doing something similar now. Recently, used large-scale RNA-seq of dermal fibroblasts from 133 people from the age of 1 to 94 - https://genomebiology.biomedcentral.com/articles/10.1186/s13059-018-1599-6 - to develop a machine learning model to predict ones age solely from the RNA-seq data with a pretty low error rate, median of 4 years.
Past
Using the fruit fly olfactory system as inspiration to invent a similarity search algorithm - https://science.sciencemag.org/content/358/6364/793.full - using three lessons, feed-forward connections, expansion of the number of neurons, and a single neuron giving inhibitory feedback, the lab invented a new tool to discover similar objects within a database:

Protein-protein interaction (PPI) networks still have major problems outstanding comprehensiveness. The cell is a complex soup and often current tools miss out on a lot of components of a given pathway. The lab developed an algorithm that minimizes the distance between the signal and target of a given pathway, osmotic stress response in yeast for the paper, to ingest current PPI data and search for potentially new interactions - https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1002640

Kaech Lab
Production of memory T-cells.Recent
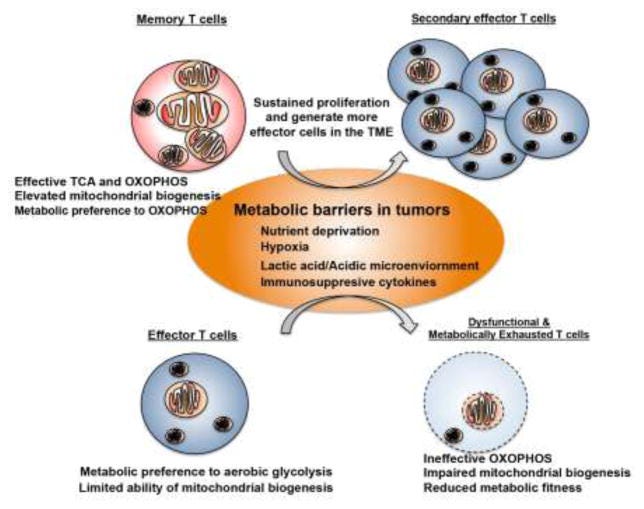
T-cells continually stimulated overtime lose their vitality, a little like playing a lot video games or going to Las Vegas. The Keach Lab discovered that a small subset of killer T-cells that express SIRPα are actually functional even after the overall population is exhausted - https://www.nature.com/articles/s41467-019-08637-9
A great overview on a new and exciting target driving T-cell exhaustion, a really important problem to make cell therapies more useful - https://www.nature.com/articles/s41590-019-0478-y - a short review on the transcription factor TOX and its role on CD8 positive T-cells (the ones that kill things) persistence/exhaustion:
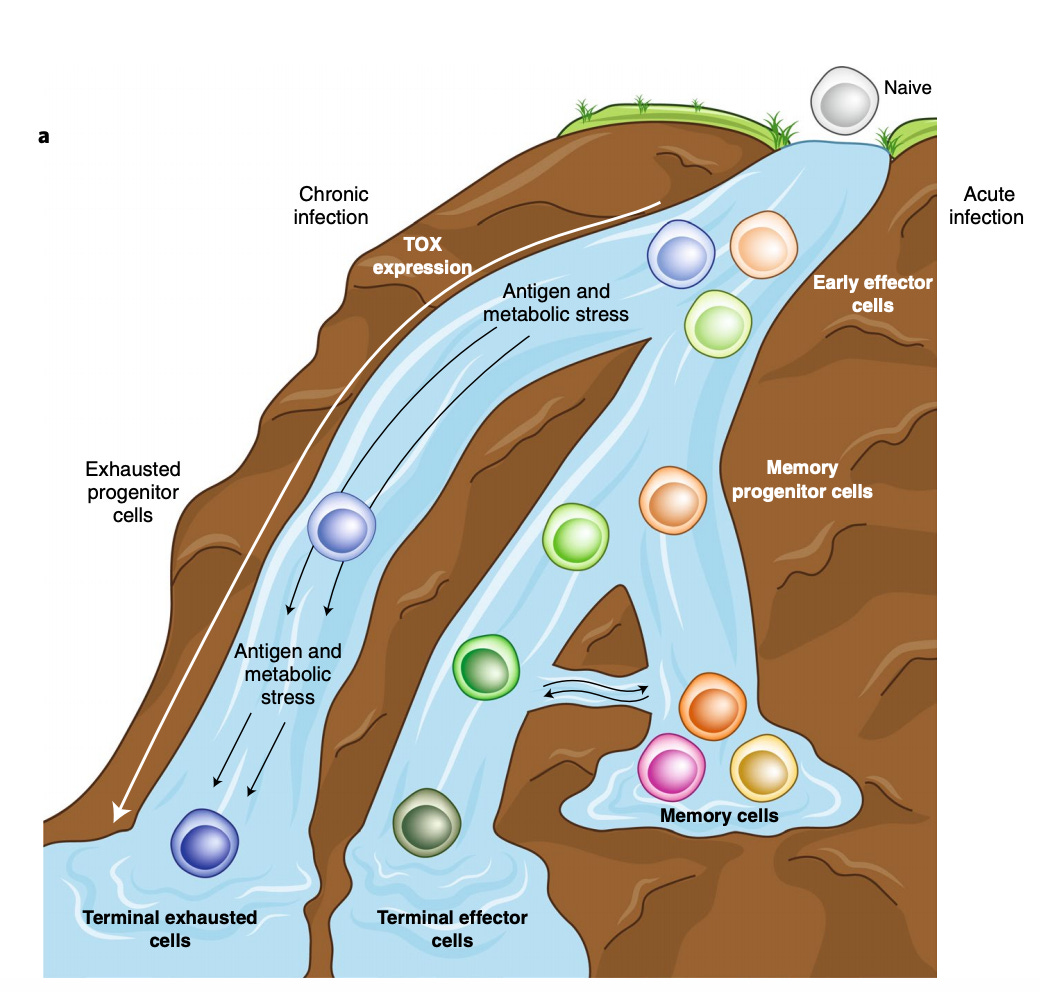
A really neat paper to connect certain cell types to the potential for a patient to respond to a drug - http://jem.rupress.org/content/215/3/877.long - simply, using tumor-associated myeloid cells and macrophages (TAM) and their secretions to create the starting point to engineer a tumor from cold to hot (non-responder to responder).
Past
Great overview on how cancer cells evade the immune system by modulating immunometabolism - https://www.ncbi.nlm.nih.gov/pubmed/?term=28458087%5BPMID%5D
Lee Lab
Bringing genetics to study nerve regeneration for spinal cord injury.Recent
Glial cells are incredible important for the brain, they are the most abundant cell type in the central nervous system and under-appreciated right now. The Lee Lab using traditional genetic screens to discover that thrombin from the liver (serine protease) regulate neurodegeneration through a mechanism via Schwann cells (glial subtype) - https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1007948
Past
Using genetics to discover factors that drive recovery from spinal cord injury; finding that p45 promotes recovery through mechanism with Fas-associated death domain (DD) adaptor (FADD) - https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0069286
Nimmerjahn Lab
Inventing new imaging tools to study the central nervous system.Recent
The Nimmerjahn Lab produces some of my favorite papers; I really enjoy microscopy research. Imaging tools (i.e. cryo-EM, lattice sheet) is going to transform biology and medicine. Luckily, I live a few blocks from the only lattice sheet microscope on the West Coast and down the hill from incredible cryo-EM labs - https://www.ncbi.nlm.nih.gov/pubmed/?term=31175843%5BPMID%5D - recently, writing a review of the current toolkit to live image the spinal cord in mice:

Past
Using two-photon and one-photon microscopy to live image cells in spinal cords in mice - https://www.nature.com/articles/ncomms11450
An interesting overview of microglia, a subtype of glia cells acting as macrophages in the brain and spinal cord -https://www.jneurosci.org/content/31/45/16064.long
Astrocytes are an incredibly unique cell types involved in blood-brain barrier formation and spinal cord injury recovery. Second to only neutrophils, astrocytes are personally to me the most interesting and ignored cell type. Doing a pretty comprehensive review of using new microscopy tools to image and augment astrocytes in vivo - https://www.ncbi.nlm.nih.gov/pubmed/?term=25665733%5BPMID%5D

Shaw Lab
Studying cell metabolism and growth.Recent
Studying a pathway that preemptively removes pre-cancerous cells, replicative crisis - https://www.nature.com/articles/s41586-019-0885-0 - to discover that telomere-activated via cGAS/STING autophagy is a key driver for the pathway.
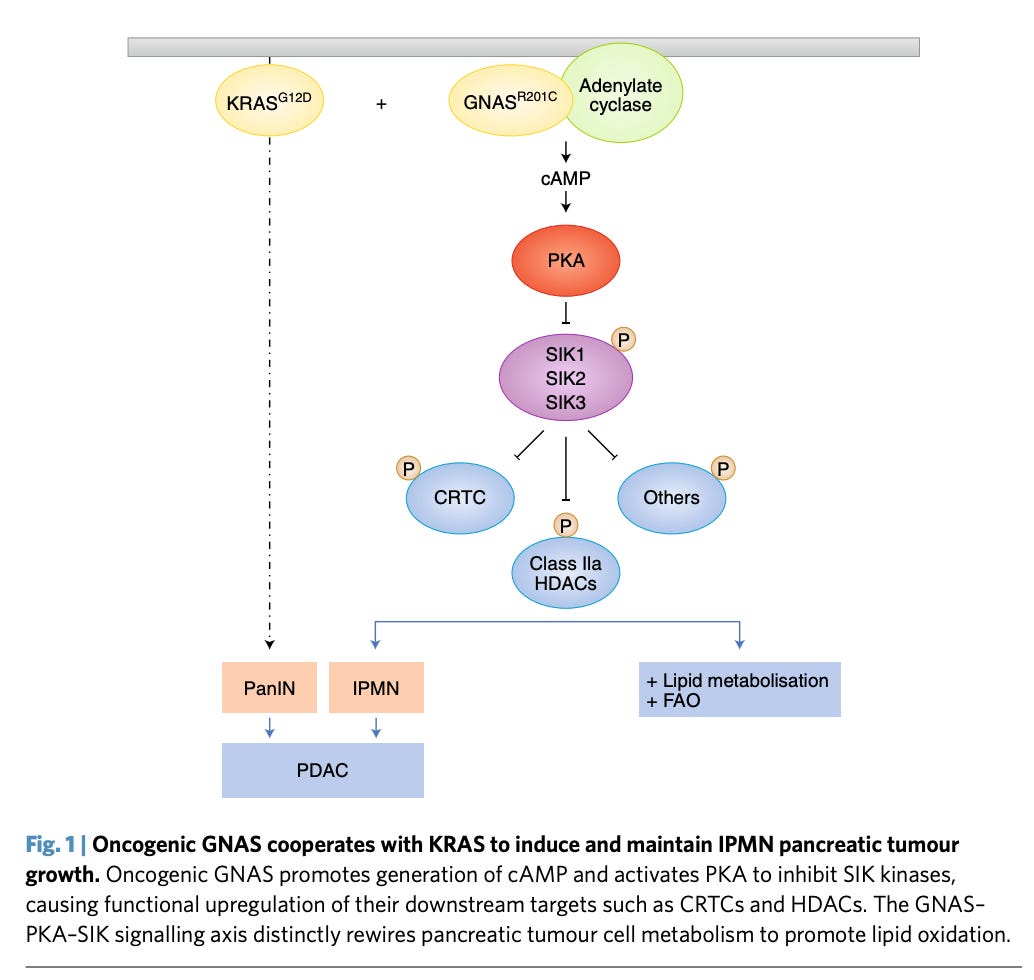
Using genetic screens to study AMPK family kinases and their role in non–small cell lung cancer (NSCLC) - https://cancerdiscovery.aacrjournals.org/content/9/11/1606.long - discovering two new kinases Sik1/3 as new targets.
A news and views on a paper from MGH discovering the role of Concurrent G-protein αs (GNAS) on inhibiting SIK kinases and driving pancreatic cancer - https://www.nature.com/articles/s41556-018-0120-5
Past
A useful review of the role of AMPK on mitochondrial health and activity - https://www.nature.com/articles/nrm.2017.95 - particularly around mitophagy, fission, and replication:


Hetzer Lab
Studying the organization of the nucleus and its role in disease.Recent
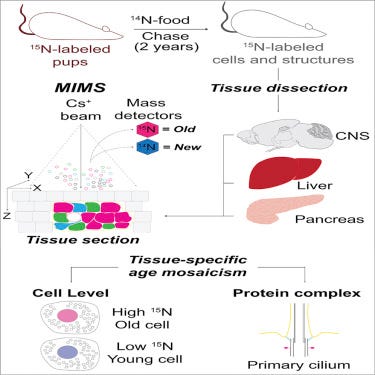
In June 2019, published a really neat paper that should have gotten more attention on the mosaicism of cell age across tissue types -https://www.cell.com/cell-metabolism/fulltext/S1550-4131(19)30250-5 - using good old fashion pulse-chase experiments and isotope microscopy, mapped out the age of cells across different tissues in mice setting the basis for further research delving deeper say into beta cells in the pancreas:
Discovering that the protein Tpr depletion dramatically increases the number of nuclear pore complexes across different cell types - http://genesdev.cshlp.org/content/32/19-20/1321.long - induced by ERK phosphorylation within the MAPK pathway.
Past
Linking the 3D genome to expression by discovering that nuclear pore complex components Nup93/153 binding superenhancer sites - http://genesdev.cshlp.org/content/30/20/2253.long - bringing them to the periphery of the nucleus, a transcriptionally repressive region.
Similar to the June 2019 paper, doing a pulse-chase experiment in rat brains to measure the lifespans of proteins particularly focused on histones and nuclear pore complexes - https://www.cell.com/cell/fulltext/S0092-8674(13)00945-8


