
Organoids
The Arlotta Lab published one of the most exciting papers in 2019 - combining single-cell sequencing with organoids to create a synthetic cerebral cortex matching the diversity found in the human brain. What was outstanding about the work was the various tools that were brought to bear to the problem of modeling the human cerebral cortex and building a method to potentially discover new medicines to treat neurodegeneration. From my personal opinion, the paper did not get the attention it deserved; a few people who are experts in organoids raved about the work but overall, the scientific and business community seemed to not appreciate how much a tour de force nature of this work.
Key experiments:
Iterating through a set of protocols to initiate cerebral cortex organoid development.
With a set protocol, using single-cell RNA sequencing along with immunohistochemistry to monitor organoid development and ensure its accuracy.
Then measure variability between organoids and between organoids and humans.
What milestones to design a business or commercial project around this invention:
The paper does a great job brining a set of tools together and laying the groundwork for a potential drug discovery platform. The next key experiment is to identify a valuable target(s) and screen a library against the organoids to show that drug screening is possible.
Afterwards, the business model becomes pretty self-evident with companies like Emulate (organ-on-chip) showing the way - either tools or internal development.
Organoids are a powerful concept without the track record yet. The ability to create synthetic models of human organs and tissues to screen for new drugs is very exciting. However, the modeling part has been lacking historically. There’s always been hype around organoids, investors lose money and patients lose out, but new inventions are being developed to make organoids more useful for drug development. This paper brings together two tools - iPSC engineering and single-cell sequencing - to design organoids that to-date best replicate the human cerebral cortex. With money from the Broad, the group used iPS cells to create organoids and single-cell sequencing to continually monitor them to ensure low variability between each organoid.
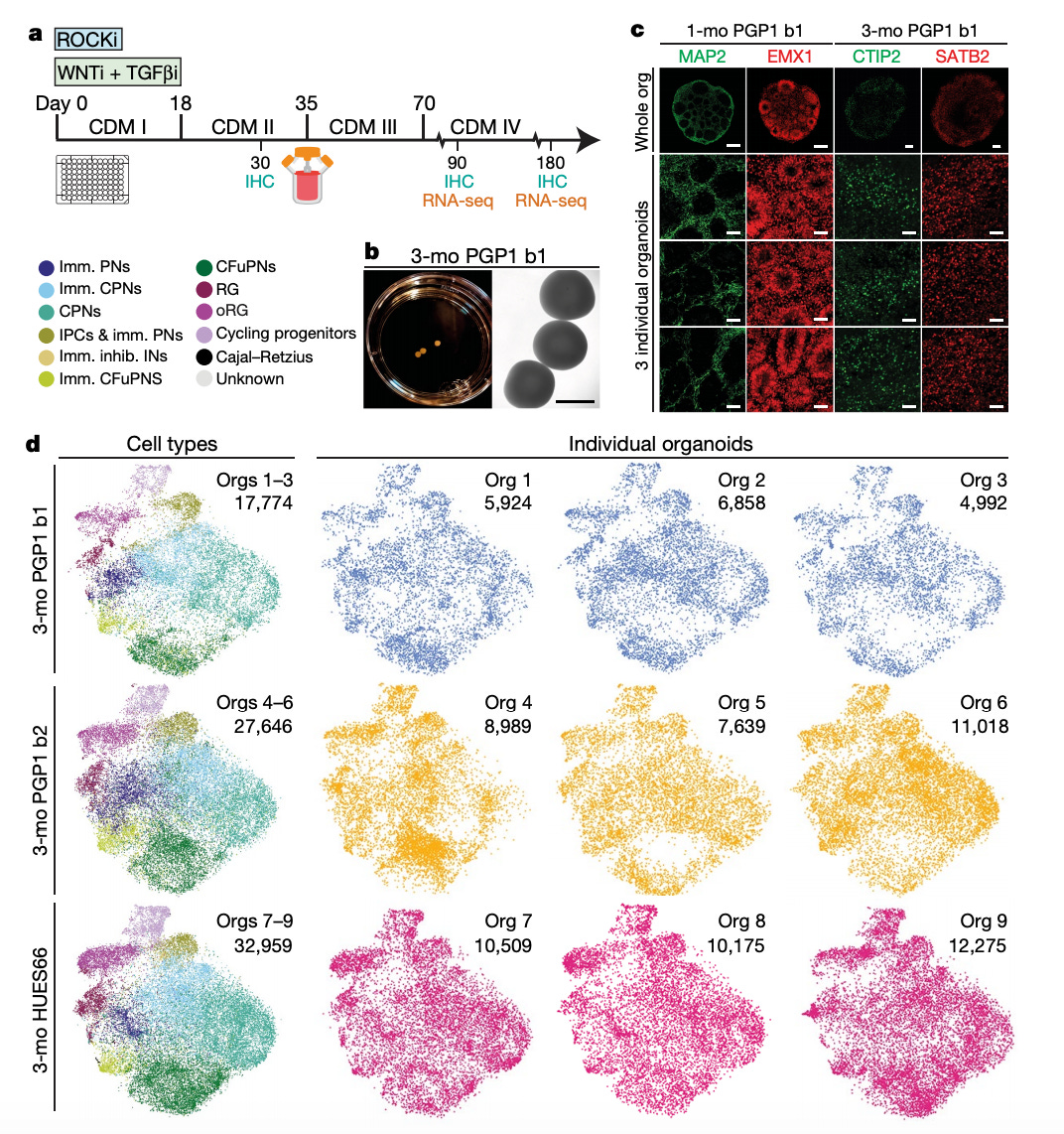
The first step was to design a protocol to produce brain organoids and validate the method. The group tested 4 different methods. Figure 1a shows the chosen protocol that uses ROCK, Wnt, and TGF-β inhibition to initiate iPSC growth/differentiation. Grown in spinner-flask bioreactors with cortical differentiation medium (CDM), immunohistochemistry and single-cell RNA sequencing were used at day 30, 90, and 180 to measure progress and consistency. The key experiment is in Figure 1d - across two iPS cell lines (PGP1, HEUS66), the group did single-cell RNA-sequencing on 9 separate organoids at 90 days constituting over 70K individual cells. On the data set, the group performed a t-SNE analysis to identify 11 distinct classes of cells (i.e. progenitor, neuron). After doing this analysis for each organoid and composing cluster graphs, the next step was to overlay each graph upon one anther to measure variability between each organoid. The group discovered high levels of consistency between organoids even from different cell lines albeit the sample size is small - n=9.

After measuring the brain organoids diversity at 3 months, the next step was to do the same at 6 months. It is nice when money is not an object in academic research. The group observed a high level of diversity in 6 month organoids when compared to 3 month ones as shown in Figure 2b. The number of cell classes expanded from 11 to 13. Using the same analysis, Figure 2a, the group observed no dramatic increase in variability between organoids (expanded to n=11).

After validating the organoid production method and its reliability, the next step was to see how similar the organoids were to real human cerebral cortexes. Using public single-cell RNA data of cerebral cortexes in human fetuses, the paper trained a random forest classifier to apply to the organoid data set to classify cells. Figure 3d, shows a map of cells types, dot size shows percentage with a class, across three organoids at 3 months. The paper did a similar experiment for organoids at 6 months (in supplementary) and discovered that their transcriptional profiles match the human fetal cerebral cortex - correlation coefficients between 0.67 and 0.80 as shown in Figure 3e.
Ultimately, the paper shows the power of using single-cell sequencing to enable consistency in organoid design and production. The paper conducted single-cell RNA sequencing across over 160K cells from 21 organoids to develop a methodology to produce models that more accurately recapitulate the cortex. The next steps are to understand how exogenous signaling at the beginning of the method or along the way can improve accuracy, get the correlation coefficients to >0.95, and show an ability to create consistent organoids from a more diverse set of cell lines.


