Axial - Inventors #14
Surveying great inventors and businesses
Axial partners with great founders and inventors. We invest in early-stage life sciences companies such as Appia Bio, Seranova Bio, Delix Therapeutics, Simcha Therapeutics, among others often when they are no more than an idea. We are fanatical about helping the rare inventor who is compelled to build their own enduring business. If you or someone you know has a great idea or company in life sciences, Axial would be excited to get to know you and possibly invest in your vision and company . We are excited to be in business with you - email us at info@axialvc.com
Inventors #14
A set of ideas and observations on inventions and discoveries in life sciences.
Immunology
The immune system and everything in it.
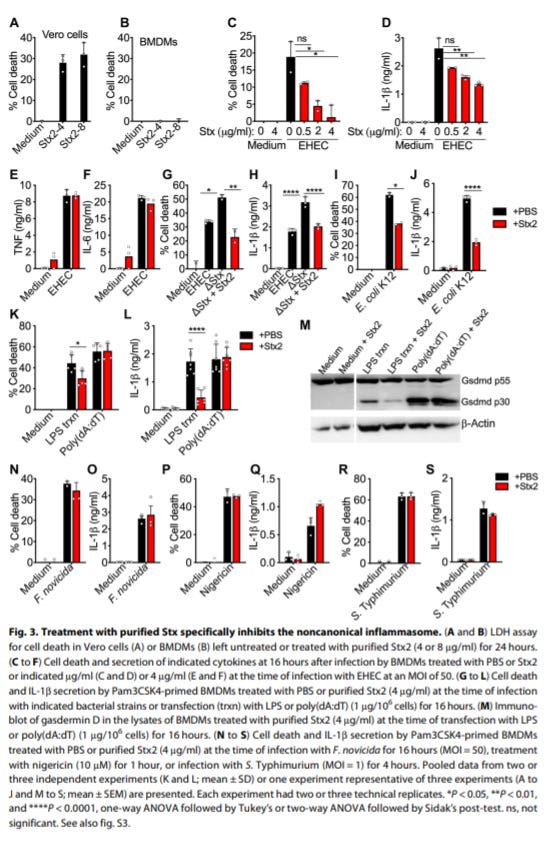
Shiga toxin suppresses noncanonical inflammasome responses to cytosolic LPS - https://immunology.sciencemag.org/content/5/53/eabc0217 - the Vanaja Lab from UConn discovered that Shiga toxin (Stx), a toxin produced by Enterohemorrhagic E. coli (EHEC; cause of colitis) and S. dysenteriae (cause of dysentery), inhibit inflammasome activity to minimize immune system detection of an infection:
Pathogens use a diverse palette of toxins to subvert the human immune system; many of these toxins actually activate inflammasomes, multi-protein complexes in the innate immune system. Inflammasomes are involved in sensing signals from pathogens, and regulating the activation of caspases and inducing an inflammatory response.
This paper did a great job at showing that a toxin, Stx, directly inhibits inflammasomes - suppressing “caspase-11–mediated activation of the cytosolic lipopolysaccharide (LPS) sensing pathway”
In the backdrop of previous work that suggested that Stx actually activates inflammasomes, which was an experimental artifact due to LPS contamination, the Vanaja Lab’s work to show that Stx inhibits inflammasomes in vivo after LPS challenge is an important step forward to understand how pathogens evade the immune system
Biochemistry and structural biology
The granddaddy of them all.
A Bump-Hole Strategy for Increased Stringency of Cell-Specific Metabolic Labeling of RNA - https://pubs.acs.org/doi/10.1021/acschembio.0c00755 - out of the Spitale Lab at UC Irvine, the group invented a tool for cell-specific RNA labeling:
The premise of the tool is centered around a bump-and-hole strategy - creating a hole in an enzyme to accommodate a bump or a larger functional group on a substrate; this concept imbues specificity between the mutant enzyme and new substrate. The group had previously generated mutant uracil phosphoribosyltransferases (mUPRT) to successfully incorporate modified 5-uracil analogs into RNA.
They found that 5-vinyluracil was a nucleobase that had minimal background integration due to steric clash within the activate site wild-type UPRT
Using their mUPRT and 5-vinyluracil, the paper showed successful selective labeling of RNA in vitro
This demonstration of a bio-orthogonal nucleoside to label RNA can be used to measure cell-specific RNA expression and open up a suite of new experiments. Next steps are to show the tool works in vivo.
Neuroscience
Roughly 20 years behind but set up to transform the concept of human.
A Chromatin Accessibility Atlas of the Developing Human Telencephalon - https://www.cell.com/cell/fulltext/S0092-8674(20)30689-9 - from the Rubenstein Lab at UCSF, a map of chromatin accessibility of the human cerebrum was created:
The group used ATAC-seq on nine regions of the human cerebrum and two parts of the prefrontal cortex sourced from fetal samples to measure chromatin accessibility
Relying on machine learning to analyze open and closed chromatin regions anchored around known promoters and enhancers, regulatory elements (RE) serving as genetic enhancers were predicted
With this dataset of over 19K predicted REs, the group discovered two variants in these regions that mapped near an autism risk gene, SLC6A1
After discovering these two variants, an CRISPRa (activation) screen was used to validate the the REs regulate SLC6A1 gene expression in neuroblastoma cells
This resource paper created a dataset of region- and age-specific REs in the human brain. This type of work can form the basis of understanding basic neurobiology as well as identifying new drug targets for diseases like Alzheimer’s and autism.
Cell biology
Cell structure and function.
Global architecture of the nucleus in single cells by DNA seqFISH+ and multiplexed immunofluorescence - https://www.biorxiv.org/content/10.1101/2020.11.29.403055v1 - the Cai Lab at Caltech imaged thousands of genetic loci in single mouse embryonic stem cells (mESCs) to connect gene expression in the nucleus to physical location:
The group combined 2 tools to image over 3,600 loci in single cells and connect them to gene expression markers: (1) DNA and RNA seqFISH (2) immunofluorescence of histone modifications to measure chromatin accessibility
With this multiplexed strategy the Cai Lab identified 12 nuclear zones determined by certain chromatin marks with some actually heritable for up to 4 generations - one zones characterized by the enrichment of SF3a66, a splicing factor, the nuclear lamina driven by Lamin B1 and the nucleolus enriched for Fibrillarin, and histone acetylation and methylation determining other zones
Moreover, the study found active genetic loci characterized by nascent transcripts were more likely found between these 12 nuclear zones
Overall, this work brings a higher resolution to the understanding of nuclear organization and shows the power of seqFISH to map the spatial features of gnomes in single cells
seqFISH can “accurately detect low copy number genes, such as transcription factors and master regulators, that cannot be detected with single cell RNAseq or immunostaining” as well as being mixed-and-matched with other imaging and sequencing methods across a wide range of cell types
Bringing new imaging modalities to the nucleus has unlocked new discoveries around how genes and proteins separate (i.e. liquid droplet-like) and how proteins like transcription factors move in 3D space to bind target regions like promoters. Imaging not only lays out the rules of nuclear organization but can help determine in vivo kinetics, which will set up more precise drug development targeting gene expression.
Genetics, genomics, and developmental biology
Heredity and variation.
Host variables confound gut microbiota studies of human disease - https://www.nature.com/articles/s41586-020-2881-9 - out of the Belkaid Lab at NIH, the group studied the impact of host variables - “body mass index (BMI), sex, age, geographical location, alcohol consumption frequency, bowel movement quality (BMQ), and dietary intake frequency of meat/eggs, dairy, vegetables, whole grain and salted snacks” - on the relationship between disease and the gut microbiome:
Relying on patient data from the American Gut Consortium, the group looked for host variables that drive microbiome diversity for individuals with inflammatory bowel disease (IBD), autism spectrum disorder (ASD), type 2 diabetes (T2D), and 16 other diseases
The study found that the frequency of alcohol consumption and bowel movements were significant indicators of microbiome variance between healthy and diseased patients
These types of studies are important to begin to realize the potential of the microbiome in human health - confounding variables especially those around individual lifestyle make determining real cause/effect relationships between the microbiome and disease nearly impossible
From this study, patients matched for their alcohol consumption and bowel movements will lead to more significant conclusions on whether changes to the microbiome is actually driving the pathology of a disease
The long-term potential is to do these types of studies at a population scale to know where the microbiome plays a significant role in a disease and where/when to intervene for a patient