Axial - Boston University #1
Analysis of exciting Boston University life sciences inventors and their inventions
The Boston University Bridge has my favorite spot to sit at and relax while jogging along the Charles River. Also, BU probably throws the best college parties in Boston. MIT is second. Harvard is close to last. In terms of scale of research programs, the ranking is flipped somewhat. Despite that, Boston University is a powerhouse in synthetic biology (even with Collins going off to MIT) already driving the formation of companies from Beta Bionics and Lattice Automation to Synlogic.

Khalil Lab
Engineering and evolving biological systems.Recent
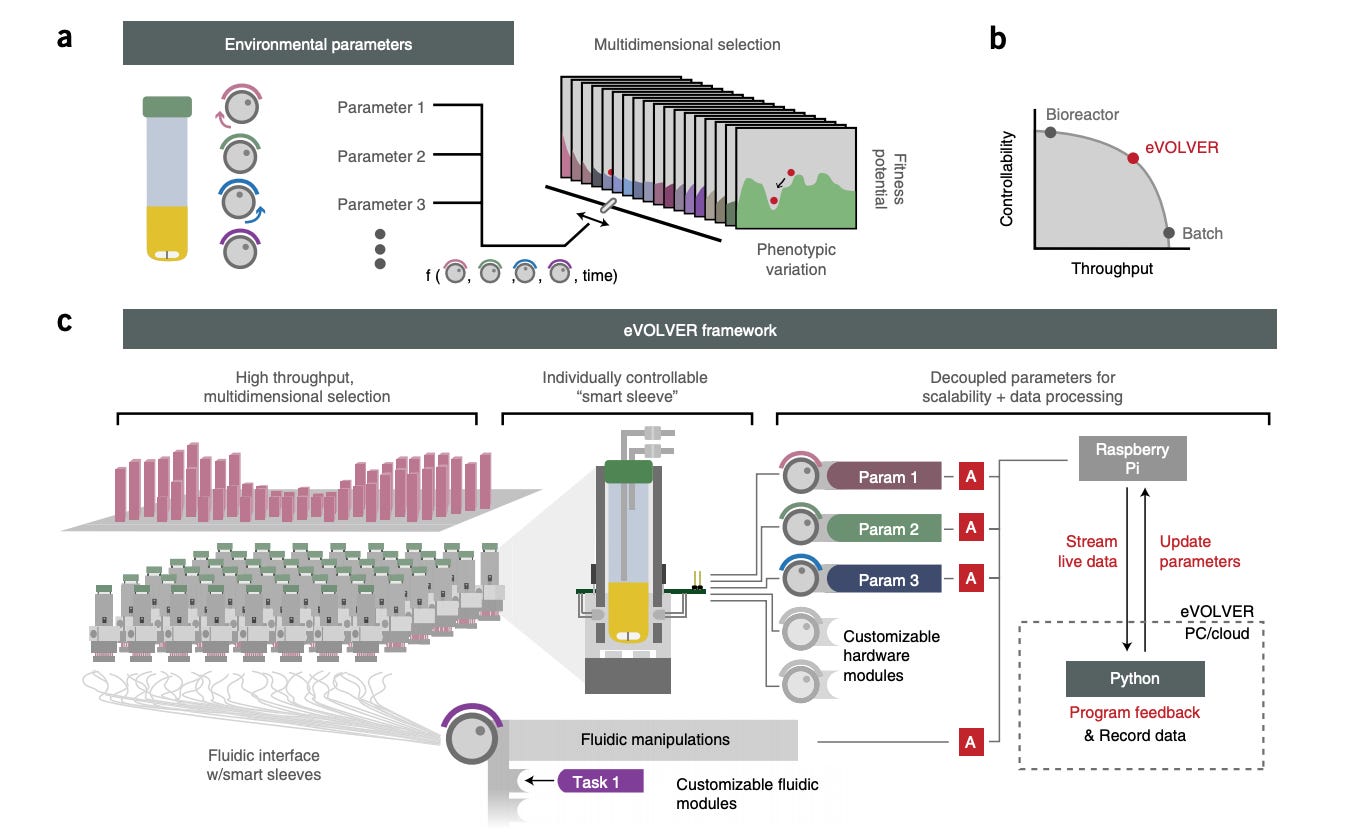
The Khalil Lab is doing incredible work in synthetic biology. Recently, the group invented a method to automate cell-culture systems called eVOLVER enabling high-throughput evolution and selection of yeast strains - https://www.bu.edu/khalillab/_papers/wong-nbt2018.pdf - effectively, the tool couples fermentation to design. Yeast knockouts across a wide variety of conditions can discover strains with unique properties - certain yield at a high temperature, induction of growth and production phases. Companies like Culture are doing incredible work to bring this paradigm to market.

Predicting the effects of drug combinations not only for diseases but for individual patients is an incredible difficult problem. There are over a thousand checkpoint inhibitor trials undergoing and various proposals to combine a new therapy with an existing one to improve efficacy/tox profiles. However, knowing which combinations to move forward with upfront rather than spending the money to do the mice experiments and sometimes humans to see if the combos actually work as expected would be transformational. The Khalil lab designed a high-throughput screening method to assess the pairwise interactions of 12 anti-fungal small molecules in both C. albicans and S. cerevisiae - https://www.bu.edu/khalillab/_papers/weinstein-natcomm2018.pdf - the experiments are retrospectively simple but lay out a elegant framework to determine drug interactions within a model and measure synergy between models:

Past
From a business perspective and investing, creating products and businesses on tools that enable control down the Central Dogma is pretty obvious. On the back of decades of work around genetics and sequencing, genome engineering has gain substantial traction. Another level up, epigenetics (i.e. the modification of the genome; for example DNA methylation or histone acetylation) is still being elucidated providing an opportunity to engineer the epigenome and built a series of great businesses. The Khalil Lab composed an important overview of the existing toolkit for epigenome engineering and the potential future - https://www.bu.edu/khalillab/_papers/park-gb2016.pdf - the cause/effect relationships of chromatin marks still need to be mapped out and editing tools need more precision:

Wong Lab
Using synthetic biology to cure cancer.Recent
The ability to engineer multi-domain proteins is an incredible opportunity particular to improve the efficacy and speed up the design process of cell/gene therapies. Serotiny is doing the best work here to build the largest libraries of these proteins. Generating the actual effector of a cell therapy is a major bottleneck for the introduction of new curative medicines; so for CAR-T, it’s the chimeric antigen receptor (CAR). The Wong Lab recently did a series of elegant experiments to build up a CAR and validate for a new cancer target, Axl - https://wilsonwonglab.org/wp-content/uploads/2019/05/Cho-2018-2.pdf

The Wong Lab is doing world-class work in synthetic biology so a recent review on the synthetic biology toolkit (present day and where it can improve) for immunotherapies is a pleasure to read - https://wilsonwonglab.org/wp-content/uploads/2019/05/Brenner-2018.pdf - the three main levers are receptor engineering, safety/logic control, and pure efficacy (i.e. binding a target). Designing an antibody is mainly focused on efficacy - does it bind the antigen or not. Whereas, CAR proteins and other cell therapy modalities are a little different, which creates an opportunity for a new company, where binding is important as well as induction of a cellular response and overall control:

Creating a CAR protein to recognize multiple antigens and induce similar responses calling it split, universal, and programmable (SUPRA) CARs - https://wilsonwonglab.org/wp-content/uploads/2019/05/Cho-2018.pdf - effectively using a leucine zipper to be able to combine one CAR signaling domain with multiple effectors showing activity in cancer mouse models.
Past
Overview of the engineering of TCR and CAR-T cell therapies - https://wilsonwonglab.org/wp-content/uploads/2019/05/chakravarti-2015.pdf

Invented a method called Boolean logic and arithmetic through DNA excision (BLADE) to create genetic circuits in mammalian cells (human embryonic kidney and Jurkat T) - https://wilsonwonglab.org/wp-content/uploads/2019/05/Weinberg-2017.pdf - the method relies on the use of recombinases to insert or excise genetic regions to turn a gene on or off. The paper tests out each recombinase in a pairwise fashion to measure genetic activation via a GFP reporters. They had to do a lot of molecular biology but minimal optimization after transformation - the recombinase activities are relatively straightforward to predict actually minimize any potential cross-talk between circuits; CRISPR-based tools could have similar value but need a little bit more optimization. Engineering/discovering more recombinases is the key bottleneck toward using BLADE for more complex circuits.
Chen Lab
Tissue microfabrication.Recent
Invented a microfluidic chip to model blood flow with the effects of the extracellular matrix (ECM) on the vessel - https://www.nature.com/articles/s41596-019-0144-8 - the chip is manufactured with hydrogels to mimic the ECM poured in. Many organ-on-chip and microfluidic systems do a good job at measuring chemical effects but often do not account for actual physical forces things like the ECM impart:


Past
3D printing vascular patches for ischaemic disease - http://sites.bu.edu/chenlab/files/2017/10/204_Mirabella_NatureBME_2017.pdf - in rodents, showing the patches improve perfusion after ischaemia and myocardial infarction through improve arterial blood flow.
Overview of the role of mechanical forces in culture of 3D tissues and organoids - http://sites.bu.edu/chenlab/files/2017/10/195_Eyckmans_J-Cell-Sci_2017.pdf - many organoid models are useful to recapitulate organs/tissues at the earliest stages of development but have trouble modeling mature states. The latter are characterized by a fully formed extracellular matrix (ECM) that generates most of the mechanical forces cells experience:


Review on the toolkit to measure mechanical forces on cells - http://sites.bu.edu/chenlab/files/2014/01/158_Stapleton_CellCommuAdhes_2014.pdf - the hard part is figuring out which cell adhesion receptors in what context has a significant effect on the actin cytoskeleton and microtubules:


Boas Lab
Bringing photonics to neuroscience.Recent
Most MRI data sets are analyzed according to the Davis Model. In short, the model uses brain blood flow and oxygen consumption during normal breathing as the standard to measure changes blood oxygen level dependent (BOLD) signal for a patient. It’s a pretty simple model requiring a few differential equations. The Boas Lab studied a particular variable within the Davis Model, β - https://onlinelibrary.wiley.com/doi/abs/10.1002/mrm.27660 - the variable was hard coded for 1.5 (derived from Monte Carlo sampling); however, the Boas Lab used 2-photon microscopy to cross-validate any MRI data in mice to actually figure out the β variable is closer to 1 in vein and closer to 2 in small vessels/channels. This type of work will improve MRI data sets, but serves as a very important lesson to use new data sets to improve existing models whether for MRI, small molecular libraries, and anything similar.
Past
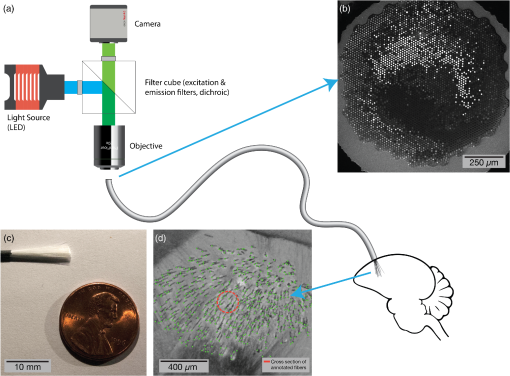
Neural probes especially within deeper regions of the brain have the potential to disrupt surrounding tissues. Moreover, there is a need to improve the accuracy of bulk measurements of neural populations. The Boas Lab modeled out the effects of thousands of optical microfibers and their distribution/density on the effects they have on neurons - https://www.spiedigitallibrary.org/journals/Neurophotonics/volume-5/issue-04/045009/Extracting-individual-neural-activity-recorded-through-splayed-optical-microfibers/10.1117/1.NPh.5.4.045009.full?SSO=1 - depending on the brain region, the group figured out the optimal fiber densities to excite neurons but figure out how much a single neuron is likely to generate a signal across multiple fibers - this allows measurements from multiple fibers to actually be useful to deduce the action potentials of individual neurons: